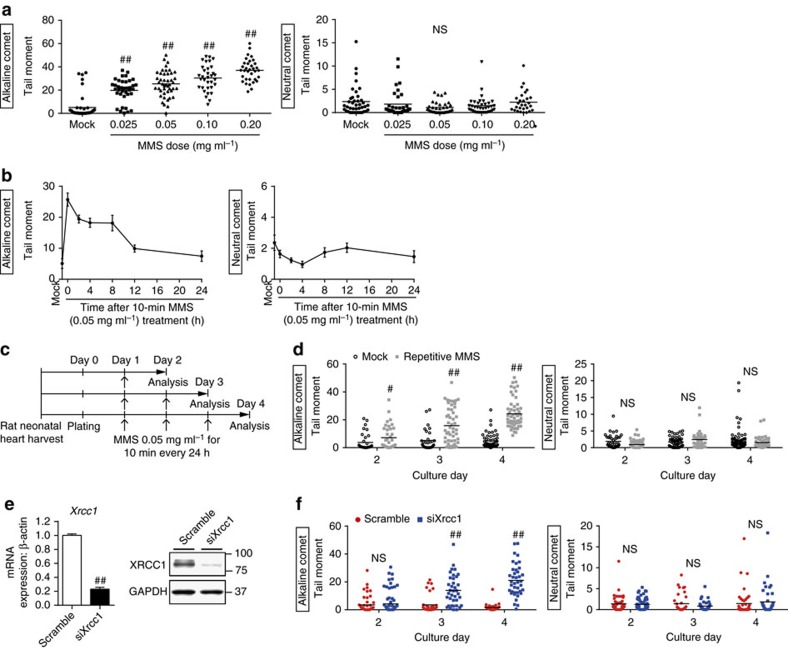
Figure 3. Generation of an in vitro model of cardiomyocytes with SSB accumulation.
(a) Neonatal rat cardiomyocytes (NRCMs) were treated with MMS at the indicated concentration for 10 min and the DNA damage was analysed by comet assay (Alkaline comet: n=42, 37, 45, 33, 34; Neutral comet: n=40, 35, 35, 37, 29 at each concentration, respectively). Statistical significance was determined by Steel–Dwass test. ##P<0.01 versus Mock. (b) NRCMs were treated with MMS (0.05 mg ml−1 for 10 min) and the DNA damage was analysed by comet assay at the indicated time point (Alkaline comet: n=41, 21, 31, 29, 16, 81, 30; Neutral comet: n=40, 36, 41, 34, 35, 41, 56 at each time point, respectively). (c) Schedule for repetitive MMS treatment. (d) NRCMs were subjected to repetitive MMS treatment as described in c and the DNA damage was analysed by comet assay (Alkaline comet: n=35, 37, 32, 49, 52, 54; Neutral comet: n=47, 54, 55, 38, 68, 45 at each time point, respectively). Statistical significance was determined by Mann-Whitney U test. #P<0.05 and ##P<0.01 versus Mock at each time point. (e,f) NRCMs were transfected with siRNA against Xrcc1 (siXrcc1) or scrambled oligonucleotide (Scramble) as a control. Knockdown efficiency of siRNA against Xrcc1 was examined by real-time PCR and western blotting (e, n=6, 8, technical duplicates). Time-dependent changes of DNA damage after the knockdown of Xrcc1 was assessed by comet assay (f, Alkaline comet: n=52, 76, 38, 37, 34, 39; Neutral comet: n=52, 65, 37, 36, 45, 41 at each time point, respectively). Statistical significance was determined by Mann–Whitney U-test for (e,f) ##P<0.01 versus Scramble at each time point. Column and error bars show mean and s.e.m., respectively.