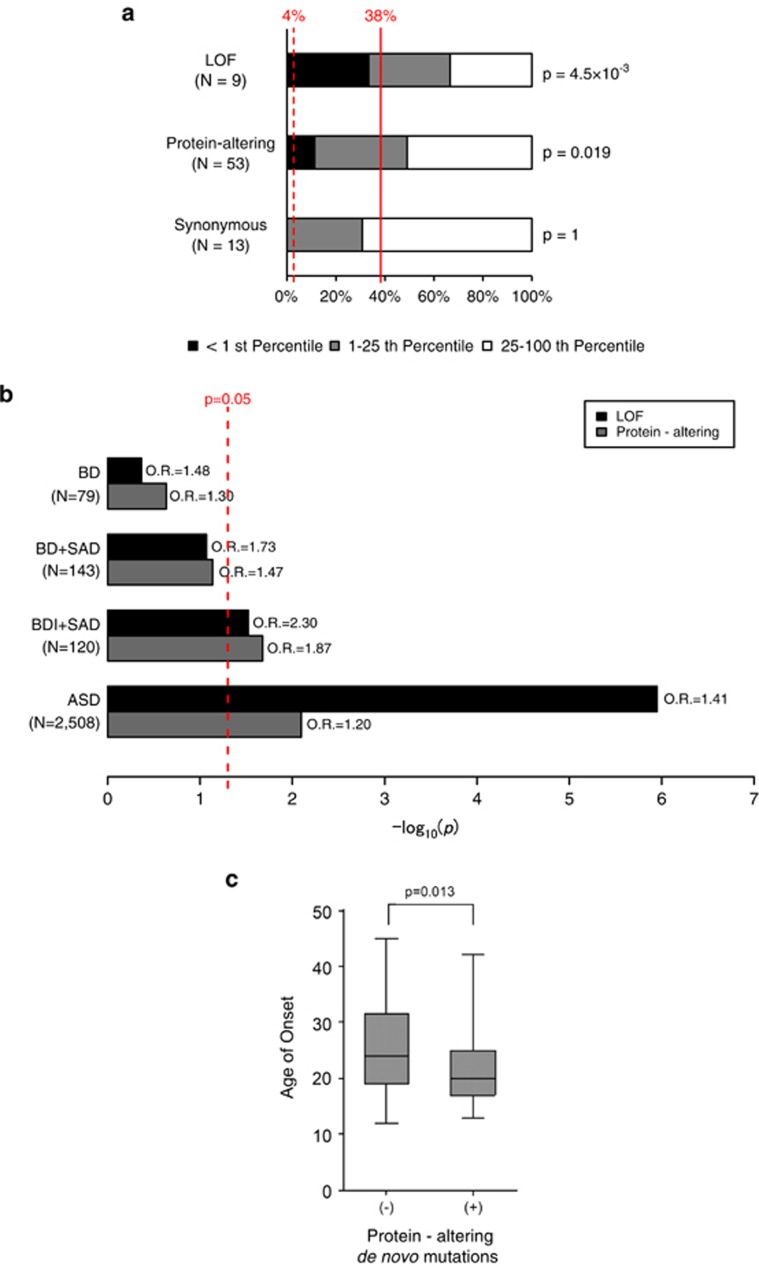
Figure 1.
Roles of de novo loss-of-function (LOF) and protein-altering mutations in bipolar disorder. (a) Proportion of the genes hit by different types of de novo mutations according to their gene intolerance. Gene intolerance to protein-altering variants in general population was assessed by using Residual Variation Intolerance Score (RVIS).38 Black, <1st percentile of intolerant genes; gray, 1–25th percentiles of intolerant genes; white, rest of the genes (25–100th percentiles). Dashed and solid red lines indicate expected proportion for the first percentile and the first quartile of intolerant genes considering gene sizes (4 and 38%, see Materials and Methods for detailed procedures). Enrichment P-values for the first percentile of intolerant genes calculated by one-tailed binominal tests are shown on the right side of the bars. (b) Enrichment analyses of de novo LOF and protein-altering mutations in case groups. Bars indicate statistical significance (log10 P-values) for enrichment of de novo LOF (black) and protein-altering (gray) mutations in each disease group. Enrichment was evaluated by comparing the numbers of de novo LOF or protein-altering mutations and synonymous mutations between each disease group, and controls (1911 unaffected siblings in Iossifov et al.20) with one-tailed Fisher's exact test. Data for autism spectrum disorder (ASD) were shown as a reference.20 Red dashed line indicates P=0.05. (c) Box plots of age of onset for bipolar disorder (BD) probands with or without protein-altering de novo mutations. Average ages of onset between the two groups were compared by two-tailed Student's t-test. Box plots of median values with hinges at the 25th and 75th percentiles and whiskers extending to the highest and lowest values are shown. BDI, bipolar I disorder; OR, odds ratio; SAD, schizoaffective disorder.