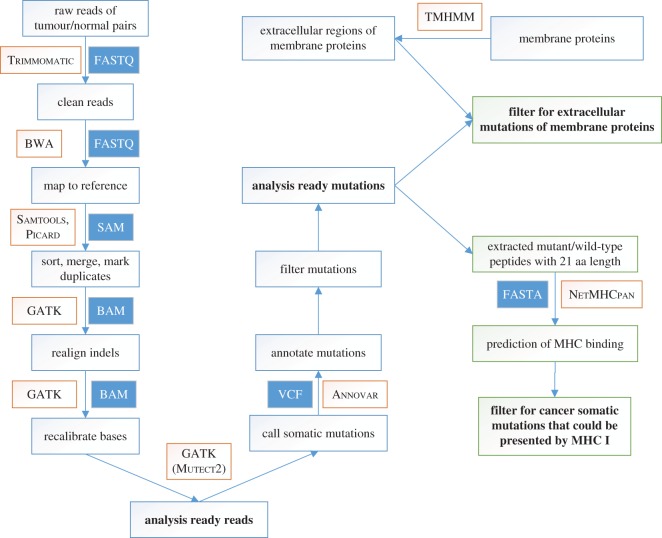
Figure 2.
The software pipeline of TSNAD. The pipeline performs best practices for somatic SNVs and INDELs in whole-genome/exome sequence with GATK. Then, we extracted the extracellular mutations of membrane proteins according to the protein topology, and invoked NetMHCpan to predict the binding information of mutant peptides to class I MHC molecules.