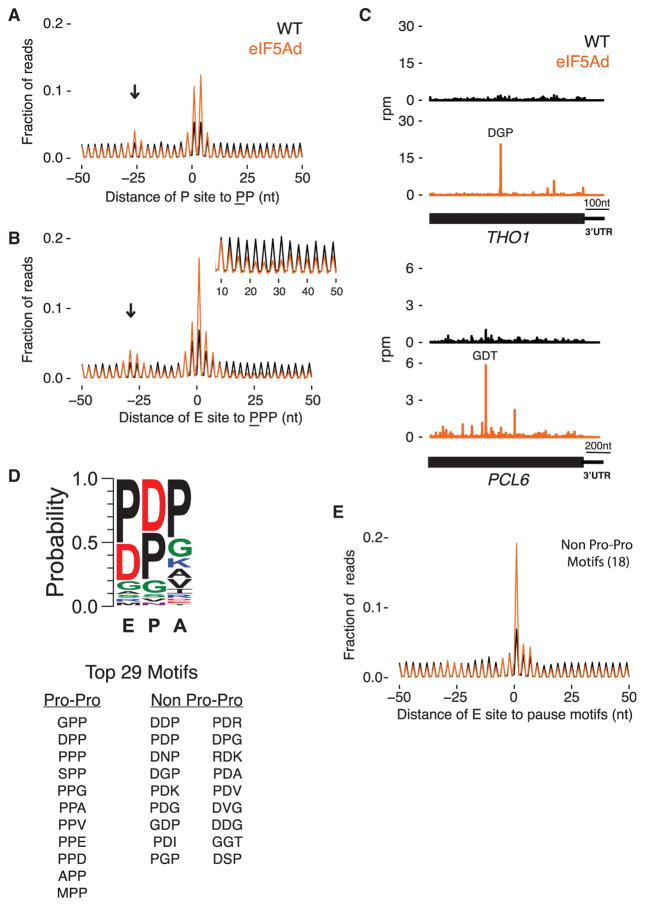
Figure 2. eIF5A Alleviates Ribosome Pausing at More Than Poly-Pro Motifs.
(A) Average ribosome occupancy centered at diproline motifs with the underlined Pro in the P site of the ribosome. Excluding motifs in genes with less than 64 reads per dataset in ORFs, 5,920 and 5,939 positions are averaged in WT and eIF5Ad, respectively. Arrow indicates stacked ribosomes.
(B) Similar to (A), average plot centered at triproline motifs with underlined Pro in the E site of the ribosome. Inset: close-up view of the ribosome occupancy 10–50 nt downstream of the triproline motif.
(C) Ribosome footprints on THO1 and PCL6 genes in WT.1 (black) and eIF5Ad.1 (orange) cells. Motifs of highest ribosome pausing are denoted. Rpm, reads per million.
(D) Peptide motif associated with ribosome pausing in eIF5Ad cells, using motifs with pause score greater than 10 and weighted by pause score ratio of eIF5Ad/WT. Top 29 motifs are listed.
(E) Average ribosome occupancy centered at 5,478 pause sites that match the 18 non-Pro-Pro pausing motifs identified in eIF5Ad cells. See also Figure S3.