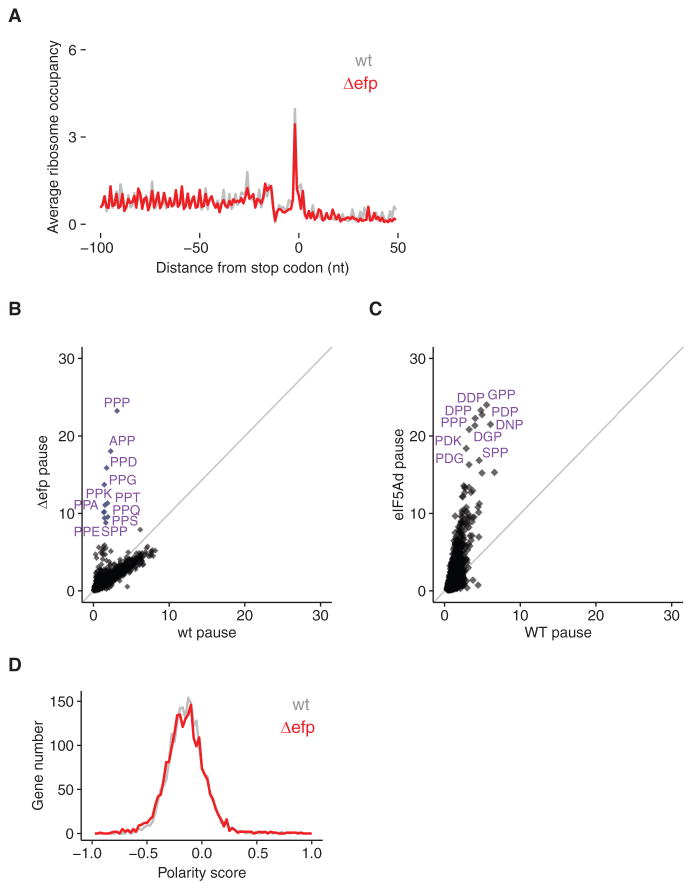
Figure 7. eIF5A Has a More General Role Than EFP.
(A) Metagene analysis of translation termination for WT and Δefp E. coli cells shows neither stacked ribosomes (~30 nt upstream of stop codons) nor 3′ UTR ribosomes.
(B) Comparison of tripeptide pausing in WT and Δefp E. coli cells. Pause scores of tripeptide motifs are plotted using E. coli WT and Δefp datasets (Woolstenhulme et al., 2015). Motifs with fewer than 30 occurrences in E. coli transcriptome are excluded from the analysis. Each dot represents one tripeptide motif; 6,018 motifs are included. Motifs with pause score higher than 8 are labeled. The diagonal line indicates the distribution expected for no enrichment.
(C) Comparison of tripeptide pausing in WT and eIF5Ad cells. Pause scores of 6,022 tripeptide motifs (Table S1) are plotted for WT and eIF5Ad cells, for motifs with more than 100 occurrences in yeast transcriptome. Ten tripeptide motifs with highest pause scores upon eIF5A depletion are labeled. The diagonal line indicates the distribution expected for no enrichment.
(D) Distributions of polarity scores for 2,186 genes from WT and Δefp E. coli cells are plotted, showing no significant difference. See also Figures S2 and S3.