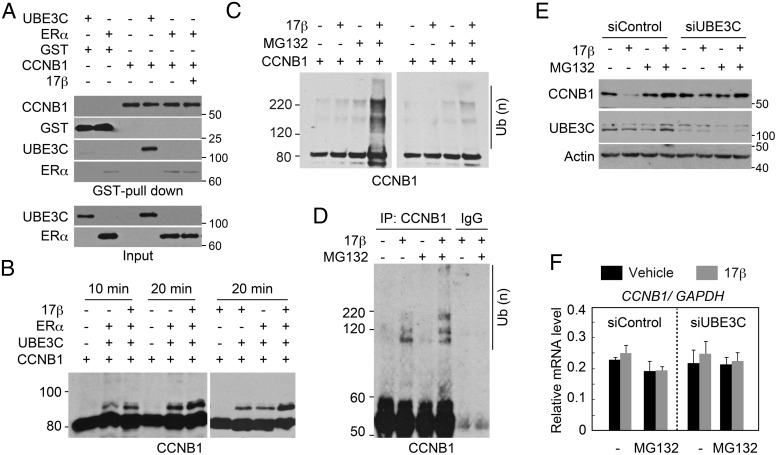
Figure 4. Estrogen enhances UBE3C-mediated ubiquitination and degradation of CCNB1.
A, Interaction of UBE3C with CCNB1 in vitro. The indicated recombinant proteins and 17β-estradiol were mixed and subjected to GST pull-down. Protein interactions were evaluated by immunoblotting with the indicated antibodies. Inputs (0.5%) were also loaded. Note that the faint amount of ERα detected in the GST-CCNB1 pull-downs is likely nonspecific, because a similar amount of ERα was detected in the control GST pull-down. B, ERα and 17β-estradiol enhance the ubiquitination of CCNB1 by UBE3C in vitro. Ubiquitin, E1, UBE2D3, and GST-CCNB1-His were incubated for the indicated time with or without FLAG-UBE3C, FLAG-ERα, and 17β-estradiol. The reactions were immunoblotted with anti-CCNB1 antibody. C, UBE3C from cells treated with estrogen is capable of ubiquitinating CCNB1 in vitro. Anti-UBE3C immunoprecipitates from mitotic MCF-7 cells treated with or without 17β-estradiol for 2 hours were mixed with ubiquitin, E1, UBE2D3, and GST-CCNB1-His and left untreated (right panel) or incubated (left panel) with ATP. MG132 was added to the cell lysate and the reaction where indicated. The reaction was immunoblotted with anti-CCNB1 antibody. D, Estrogen enhances the ubiquitination of CCNB1 in vivo. MCF-7 cells were treated with nocodazole for 24 hours. MG132 and 17β-estradiol were added as indicated during the last 4 hours of the incubation. Cell lysates were immunoprecipitated with anti-CCNB1 antibody or control IgG under denaturing conditions and subjected to immunoblotting with an anti-CCNB1 antibody. E, UBE3C-dependent destabilization of CCNB1 by estrogen during mitosis. MCF-7 cells transfected with the indicated siRNAs were treated with nocodazole for 24 hours. MG132 and 17β-estradiol were added as indicated during the last 4 hours of the incubation. Steady-state levels of CCNB1, UBE3C, and actin were analyzed by immunoblot. F, CCNB1 mRNA expression levels in cells treated as in E were analyzed by qPCR. Relative ratios normalized to GAPDH mRNA expression are shown.