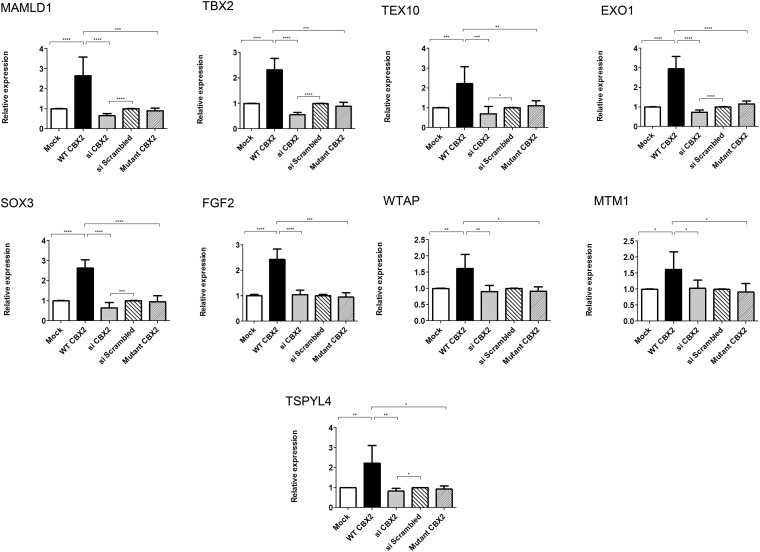
Figure 4. Genes up-regulated in a CBX2-dependent manner.
qRT-PCR quantification of mRNA levels for putative CBX2 targets that are up-regulated by CBX2. NT2–D1 cells transfected with empty vector (mock, white) or WT CBX2 (black) or mutant CBX2 (narrow lines) or scrambled siRNA (wide lines) or siRNA against CBX2 (gray). Relative expression levels of target genes were determined by qRT-PCR after normalization to cyclophillin as an endogenous control. After CBX2 overexpression (black), the relative expression of MAMLD1, TEX10, EXO1, SOX3, TBX2, FGF2, WTAP, MTM1, and TSPYL4 was up-regulated compared with cells transfected with empty vector (white). CBX2 knockdown using siRNA (gray) reduced the relative expression of MAMLD1, TEX10, EXO1, SOX3, TBX2, and TSPYL4 while not significantly affecting the expression of FGF2, WTAP, and MTM1, compared with scrambled siRNA (wide lines). Introducing the mutated forms (P98L and R443P) of CBX2 (narrow lines) in equimolar amounts into the cells showed no effect on the expression of all the target genes. The data in all graphs are the average of 3 independent experiments, error bars represent SD, and values are expressed as relative to control = 1. ****, P < .0001; ***, P < .001; **, P < .01; *, P < .05.