Abstract
Differential diagnosis and therapy of heterogeneous breast tumors poses a major clinical challenge. To address the need for a comprehensive, non-invasive strategy to define the molecular and functional profiles of tumors in vivo, we investigated a novel combination of metabolic positron emission tomography (PET) and diffusion-weighted (DW) magnetic resonance imaging (MRI) in the polyoma virus middle T transgenic mouse model of breast cancer. The implementation of a voxelwise analysis for the clustering of intra- and intertumoral heterogeneity in this model resulted in a multiparametric profile based on [18F]FDG-PET and DW-MRI which identified 3 distinct tumor phenotypes in vivo, including solid acinar and solid nodular malignancies as well as cystic hyperplasia. To evaluate the feasibility of this approach for clinical use, we examined estrogen receptor-positive (ER+) and progesterone receptor-positive (PR+) breast tumors from 5 patient cases using DW-MRI and [18F]FDG-PET in a simultaneous PET/MRI system. The post-surgical in vivo PET/MRI data was correlated to whole-slide histology using the latter traditional diagnostic standard to define phenotype. By this approach, we showed how molecular, structural (microscopic, anatomic) and functional information could be simultaneously obtained non-invasively to identify precancerous and malignant subtypes within heterogeneous tumors. Combined with an automatized analysis, our results suggest that multiparametric molecular and functional imaging may be capable of providing comprehensive tumor profiling for non-invasive cancer diagnostics.
Keywords: breast cancer, in vivo phenotyping, intratumoral heterogeneity, multiparametric imaging, PET/MRI, Gaussian mixture model
Introduction
Beyond individual variances between different tumors (intertumoral heterogeneity), intratumoral heterogeneity has become a major focus in oncology (1–4). Defined by the appearance of different histological, genetic and molecular subpopulations within single tumors, intratumoral heterogeneity creates a new challenge in cancer classification and drives neoplastic progression and therapeutic resistance (5–7). To date, 50–80 % of malignant breast cancers do not display sufficient characteristics of any special histological type, but sometimes show high grades of intratumoral heterogeneity (1,2) and are subsequently classified as “invasive breast carcinoma of no special type” (IBC-NST, Supplemental Table I contains a list of abbreviations) (8).
Pathologists therefore analyze tissue sections from different regions of the tumor, reporting the highest observed grade (9). However, tumor classification and subsequent diagnosis in the clinic remain mostly based on small tissue samples that represent a single point of information rather than the status of the whole lesion.
Molecular imaging modalities, such as positron emission tomography (PET) and magnetic resonance imaging (MRI), have the potential to provide whole-lesion information non-invasively. PET shows great benefits in staging and therapy response evaluation of lung cancer and lymphoma (10–12) but has not been transferred into routine clinical practice for breast cancer patients.
Routinely performed PET imaging analyses of tumors are based on mean or hot spot values of tracer uptake, such as maximal standardized uptake value (SUVmax) or “metabolic active tumor region” evaluation (13,14). Wahl et al. described the beneficial value of the “PET response criteria in solid tumors” (PERCIST) and discussed the great advantages of hot spot analysis procedures over the mean value analysis (15). However, all of these procedures neglect a substantial part of the information, as only a small fraction of the tumor is considered. More sophisticated approaches such as radiomics, extracting large numbers of quantitative imaging features for tissue characterization (16,17) or other types of voxelwise analyses generating parametric maps of functional parameters (18,19) are rarely used in clinical routine PET.
We hypothesized, that hybrid PET/MRI can provide a clinically relevant tumor profiling and phenotyping approach of breast cancer. Thus, in this study, clinically applied PET and MRI methods using [18F]Fluorodeoxyglucose ([18F]FDG)-PET and diffusion-weighted (DW)-MRI were combined with a unique approach of voxelwise analysis, which identified different phenotypes within single tumors. To mimic the heterogeneous nature of breast cancer, polyoma virus middle T transgenic (tg(PyV-mT)) mice, that develop malignant tumors, passing through distinct stages of tumor progression, from atypical hyperplasia to mammary intraepithelial neoplasia and invasive carcinoma (IC) (20,21), were investigated. Furthermore the clinical potential of the method was proven, including a small cohort of 5 IBC-NST patients. Our clustering approach provides a measure for the number of clusters and corresponding thresholds for the subsequent identification of tumor phenotypes and allows an easy translation to other oncological entities.
Materials & Methods
Mice
All experiments were performed with female tg(PyV-mT) mice on a C57BL/6 background. All animals were housed under standardized environmental conditions with free access to food and water. The tg(PyV-mT) mice developed an average of 5 palpable tumors, composed of a mixture of glandular and cystic dysplasias and ICs (21,22).
Preclinical in vivo imaging
Preclinical in vivo studies were performed in a sequential PET/MRI setup using a dedicated 7 T small-animal scanner (ClinScan (7T-MRI), Bruker BioSpin GmbH, Ettlingen, Germany) and a small-animal PET scanner (Inveon dedicated PET (DPET), Siemens Healthcare, Knoxville, TN, USA). Five anesthetized animals (1–2% isoflurane evaporated in breathing air) at the age of 19 – 22 weeks, bearing together 26 tumors were measured on temperature-controlled animal beds. Apparent diffusion coefficient (ADC) MRI measurements using a half fourier-acquired single shot turbo spin echo sequence (HASTE, repetition time (TR) 5000 ms, echo time (TE) 112 ms, pixel size 0.21 × 0.21 mm2 in plane, 1 mm slice thickness, b-values 150, 600, 1000 s/mm2) and sequential [18F]FDG-PET/MRI (intravenously injection of 10±2 MBq of [18F]FDG; 50 min sleeping uptake, 10 min emission scan, 13 min transmission scan) were performed on two consecutive days. PET data was reconstructed using the ordered-subsets expectation maximization algorithm with 3D post-reconstruction (OSEM3D; Inveon Acquisition Workplace, Siemens Healthcare). The anatomical MRI was measured directly before the PET scans or the ADC measurements with a 3D T2-weighted turbo spin echo sequence (TSE; TR 2500 ms, TE 202 ms, voxel size 0.27 × 0.27 × 0.27 mm3).
Although not included into this report, 4 further PET/MRI measurements, investigating the PET tracers [68Ga]NODAGA-RGD, [11C]methionine, [11C]choline and [18F]FMISO were performed with the same animals previously within one week.
An additional group of 4 tg(PyV-mT) mice at the age of 20 weeks was measured in a sequential PET/MRI setup including anatomical and DW-MRI and [18F]FDG-PET. The mice were injected intravenously with 10±2 MBq of [18F]FDG. During the uptake time of 60 min, MRI measurements were performed, directly followed by the PET scans.
All experiments were approved by the Regierungspraesidium Tuebingen.
Preclinical ex vivo methods
Following the last in vivo measurement, the mice were sacrificed, and tumors were excised and prepared for H&E histology and/or autoradiography following standard procedures (Supplemental Material and Methods).
Clinical imaging
Five patients at the age of 66±10 years with biopsy-proven breast cancer lesions (estrogen receptor- and progesterone receptor-positive (ER+/PR+) IBC-NST, G2-G3) of ≥1.5 cm in size were examined using a 3T-PET/MR scanner (Biograph mMR, Siemens Healthcare). After 60 min of [18F]FDG uptake the simultaneous PET/MRI acquisition was performed (258±4 MBq, 20 min PET acquisition, 4.3 mm maximal achievable resolution (23)). The PET data was reconstructed using an OSEM3D algorithm, applying an MR-based attenuation map from segmented MR images provided by the vendor. For the MRI measurements, a dedicated 4-channel breast MRI coil (Siemens Healthcare), that was installed during all PET and MRI measurements, was used to acquire the following MRI sequences within 20 min: short time inversion recovery (STIR), DW echo planar imaging (EPI, TR 11000 ms, TE 76 ms, pixel size 1.8 × 1.8 mm2 in plane, 4 mm slice thickness, b-values 50, 800 s/mm2), contrast agent (Gadovist®, Bayer Vital GmbH, Leverkusen, Germany) enhanced (CE) dynamic imaging (T1 fast low-angle shot (FLASH) 3D) and post contrast agent T1 FLASH 3D fat saturated (TR 8.75 ms, TE 4.21 ms, voxel size 0.9 × 0.9 × 0.9 mm3). ADC maps were calculated from the DW EPI.
All patients gave written informed consent for the purpose of anonymized evaluation and publication of their data. All reported investigations were conducted in accordance with the Helsinki Declaration and with national regulations.
Clinical ex vivo methods
Post-surgery, whole slide histology of the tumors was performed and correlated to the imaging data. The excised tissues were cut in 0.5 cm slices for macroscopic analysis and fixed in formalin and embedded in paraffin. Tissue samples were taken for routine microscopic evaluation in 4 × 4 cm2 sections. Subsequently, 3 μm-H&E sections were prepared and digitized (Axioscope, Leica Microsystems, Wetzlar, Germany).
Data Analysis
Inveon Research Workplace (IRW, Siemens Healthcare) was used for in vivo image analysis. The corresponding preclinical PET and MR images were fused and coregistered employing capillaries filled with the tracer, that were placed on the animal beds as markers for the PET and MRI data fusion. The simultaneously acquired clinical data was coregistered by the scanner software. Volumes of interest (VOIs) covering the entire regions (whole breasts or lesions, respectively) were drawn on the basis of the MRI data (T2-weighted TSE for preclinical data; post contrast agent T1 FLASH for the clinical data).
Beyond the tumor-to-muscle ratios (TMR, preclinical data) and standardized uptake values (SUV, clinical data) a new voxel-based approach was developed for [18F]FDG-PET and DW-MRI using a Gaussian mixture model (GMM) in MATLAB (MathWorks, Natick, MA, USA), fitting multiple Gaussian distributions to the collective voxel data (24). For every set of data several different mixture models with up to 7 Gaussian distributions were fitted and compared using the Akaike information criterion (AIC) and Bayesian information criterion (BIC) as implemented in the MATLAB package. The intersections formed by the descending part of one Gaussian distribution and the ascending part of the following distribution were used as thresholds to separate the subpopulations. Every Gaussian distribution within the thresholds defined a distinct biomarker population. To illustrate the results, histograms of the voxel values were calculated and overlaid with the GMM fitting results.
For ADC and PET data, parametric maps were computed where each identified population was assigned to a specific color. Subsequently, for the sequentially acquired cohort multiparametric [18F]FDG/ADC maps were computed for the entire tumors.
Autoradiography correlation
To confirm the preclinical in vivo [18F]FDG results, the GMM approach was transferred to the autoradiography data of 8 tumors. Following a background correction, TMR maps were computed and pixel values were extracted. The collective data set of all tumor pixels within the 8 tumors was fitted. Using the determined thresholds, parametric autoradiography maps were computed.
Results
Conventional image analysis hampers heterogeneous tumor characterization
First results from the tg(PyV-mT) studies revealed that entire lesions (n = 26 tumors, 53 to 769 mm3 in size) could only be detected on the basis of anatomical MRI, as intratumoral [18F]FDG uptake as well as the ADC were very heterogeneous. Due to the extensive intratumoral heterogeneity, phenotypic identification of whole lesions in terms of intertumoral comparison using single value analysis, such as mean and maximal value analyses was not feasible, as both methods omitted important information (Supplemental Figure I).
Identification of intratumoral uptake populations using GMM for a voxel-based analysis in tg(PyV-mT) mice
a) Distinct [18F]FDG uptake populations correlated with phenotypic outcome
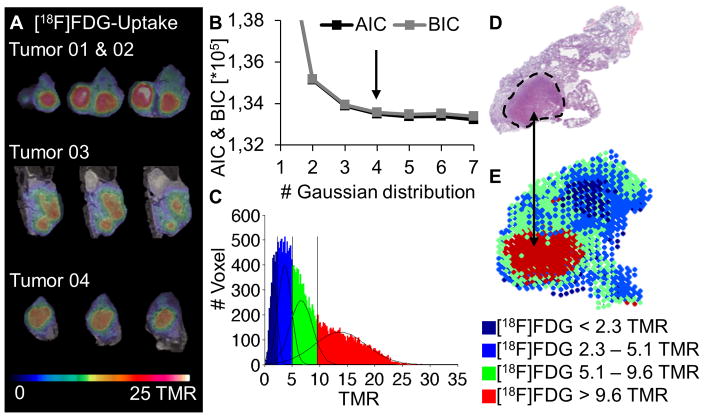
To decode the heterogeneous tumor uptake pattern that is shown for the [18F]FDG uptake of 4 representative tumors (Figure 1A), a voxel-based analysis approach for the multiparametric imaging procedure was developed. Three mice at the age of 22 weeks bearing 18 tumors were analyzed by a voxelwise calculation of the TMR for every tumor, representing the training data set which was collectively fitted using the GMM.
Figure 1.
Voxelwise analysis approach for [18F]FDG-PET: [18F]FDG-PET/MRI serial slides of 4 tumors (A). Voxel data of 18 tumors was examined. A 4 Gaussian distribution was the best fit according to AIC and BIC (B) and was applied to the summed histogram with the thresholds TMR = 2.3, 5.1 and 9.6 (C). H&E histology (D) compared to a parametric color map of the in vivo data (E).
Evaluating the AIC and BIC, a mixture of 4 uptake populations was determined as the best fit for the [18F]FDG data (Figure 1B). The intersections of the 4 distributions at TMR = 2.3, TMR = 5.1 and TMR = 9.6 represented the thresholds between the uptake populations (Figure 1C). Parametric color maps of the tumors were created by applying these thresholds on the image data (Figure 1E). Comparing to histology, these maps showed a correlation of the two populations with low [18F]FDG (blue, TMR < 5.1) uptake with cystic tumor regions, whereas solid tumor regions correlated very well with the population with the highest uptake (red, TMR > 9.6) (Figure 1D, E). However, not every Gaussian distribution directly correlated to a specific histological pattern. Due to the limited spatial resolution of the PET scanner (1.6 mm maximal achievable resolution (25)), the population with intermediate uptake, representing TMR values between 5.1 and 9.6, indicated mixed regions in which more than 1 phenotype was present within single voxels.
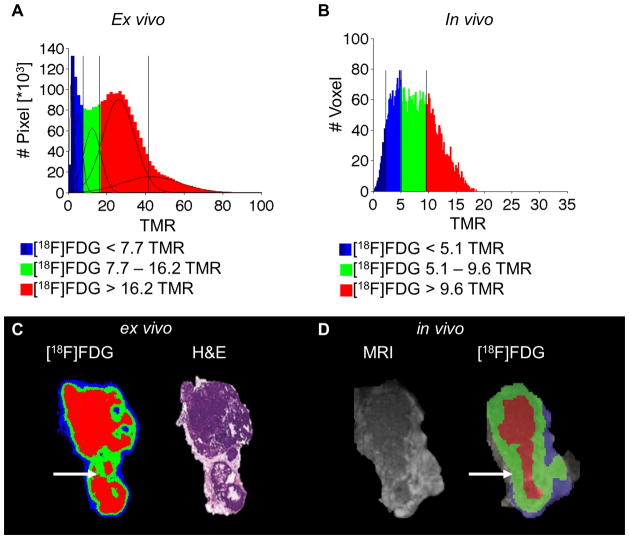
In order to confirm this phenotypic correlation, the identified thresholds from the training data set were applied to a test data set of 8 tumors (2 mice at the age of 19 weeks) without employing the GMM analysis again and correlated with ex vivo autoradiography results (Figure 2). Therefore the TMR maps of the autoradiography data of these 8 tumors were calculated and analyzed using the GMM algorithm. A total of 5 Gaussian distributions were determined as the best model ex vivo (Figure 2A). The thresholds were calculated as TMR = 3.2, 7.7, 16.2 and 41.6 and applied to the autoradiography data to compute parametric color maps. The numerical difference in the TMR thresholds between the in vivo and autoradiography measurements may be attributed to different shrinking behaviors of the tissues during the autoradiography process, including the initial freezing and subsequent drying during the exposure time as well as partial volume effects (PVE) in PET. The 41.6 threshold was not considered to be a real threshold as the 2 populations with the highest uptake values overlapped significantly and were grouped together, as a secured assignment of individual voxels to either population was not possible. As the first two blue populations (TMR < 3.2 and TMR 3.2 – 7.7) mainly corresponded to the background and spillover at the edges of the tumors, the number of dedicated tumor populations was reduced to 2 (TMR 7.7 – 16.2 and 16.2 < TMR). This result was expected due to the better spatial resolution of the autoradiography plate (0.05 mm pixel diameter) compared with the PET data, which led to a reduction of the mixed population that was observed in vivo, when more than 1 phenotype was present in a single voxel. The correlation of the in vivo histograms of single tumors (Figure 2B) and the ex vivo histograms, as well as single slices of the autoradiography (Figure 2C) with the PET data (Figure 2D), revealed a similar uptake pattern as shown for a representative tumor. The mixed areas in the PET data were mainly correlated with the population showing positive uptake by autoradiography, indicating a high amount of solid tissue in the mixed areas of the in vivo data that was verified by the corresponding H&E histology. The lower values of these regions in the in vivo data could be explained by an underestimation of the uptake due to PVE.
Figure 2.
Ex vivo correlation of the [18F]FDG data with autoradiography: the pixel values (TMR) of 8 tumors (A), and a GMM fit was applied with a result of 5 Gaussian distribution with the thresholds TMR = 3.2, 7.7,.16.2 and 41.6. The in vivo histogram of a single tumor (B) and the in vivo parametric map, revealed good correlation to the ex vivo [18F]FDG and the H&E-stained section (C) with the in vivo PET map and MRI (D). The mixed population of the in vivo data was mainly covered by the high uptake population in the ex vivo data (white arrow).
b) ADC complements the [18F]FDG pattern for population analysis
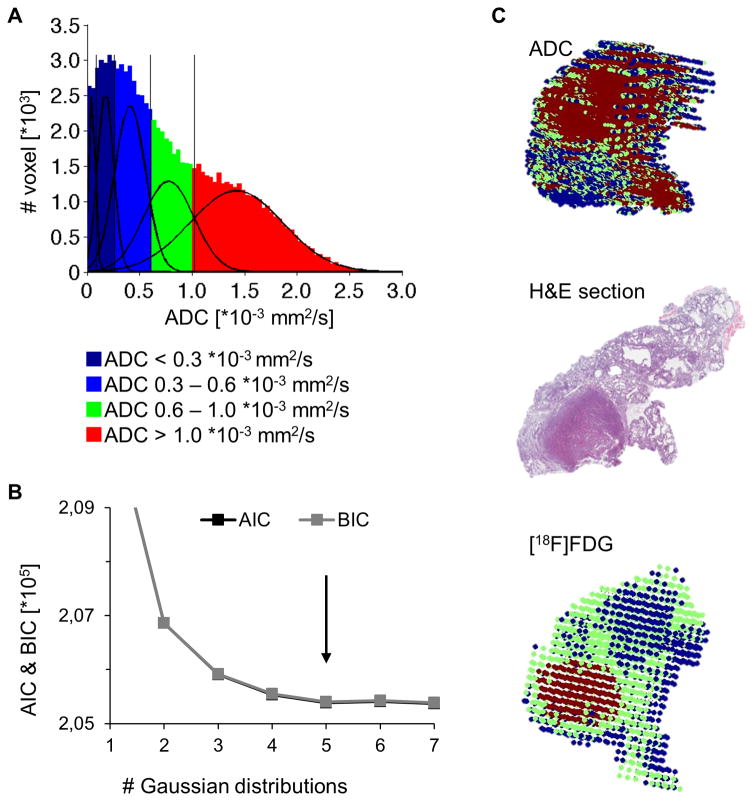
The GMM approach was also applied to the ADC data. All tumor voxels within the training data set were fitted (Figure 3A). According to the defined fitting criteria, 5 ADC populations were identified as the best model (Figure 3B). As the first and second population represented background populations (data not shown) these 2 populations could be neglected. The thresholds of the 3 remaining populations were determined as ADC = 0.6 * 10−3 mm2/s and ADC = 1.0 * 10−3 mm2/s. Mapping these populations to 3D parametric color maps and correlating these maps with the H&E histology showed a correlation of the high ADC values (ADC > 1.0 * 10−3 mm2/s) with the cystic hyperplastic regions within the lesions, while the low ADC population (ADC < 0.6 * 10−3 mm2/s) correlated with the solid regions. Similar to the [18F]FDG populations, a mixed ADC population was observed (ADC: 0.6 – 1.0 * 10−3 mm2/s), where more than 1 phenotype was present within single voxels.
Figure 3.
Voxelwise analysis approach for ADC-maps: the voxel data of 18 tumors was examined (A). A 5 Gaussian distribution was determined as the best fit according to AIC and BIC (B). 3D color maps of the [18F]FDG and ADC of a representative tumor correlated with the corresponding histology slide (C).
To compare the parameters of [18F]FDG and ADC, both classifications were applied to each tumor voxel within the training data set, to investigate the tumors’ composition according to both parameters (Figure 3C). Whereas the population with high [18F]FDG uptake (TMR > 9.6) correlated with solid regions showing high metabolic activity, ADC maps clearly revealed hyperplastic cystic regions (ADC > 1.0 *10−3 mm2/s), that could not be identified by the [18F]FDG uptake due to the low uptake values < 5.1 TMR. The combination of both methods, [18F]FDG and ADC PET/MRI, provided the opportunity to identify solid as well as cystic regions and thus, characterized the whole tumor burden within the mice without the need for tissue sections.
Multiparametric PET/MRI decodes intratumoral phenotypic heterogeneity in vivo
On the basis of these results, a multiparametric study was designed that enabled the direct correlation of the voxel data of both modalities for in vivo phenotypic clustering. Due to the sequential imaging setup, the ADC and [18F]FDG data of 4 mice at the age of 20 weeks could be directly fused for analysis. The thresholds of the training data set were directly transferred to this test data set without employing the GMM fitting again.
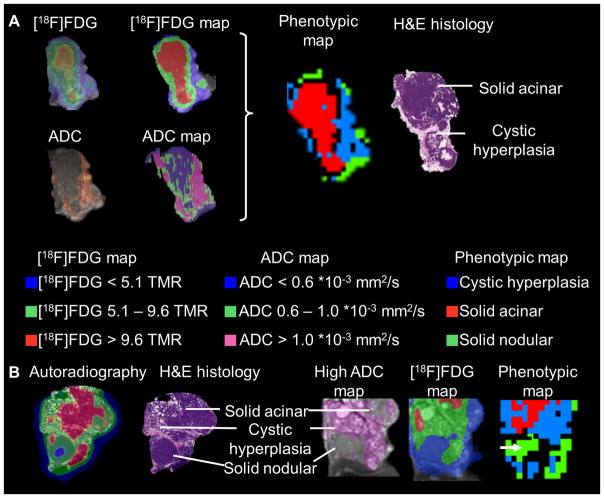
Furthermore, the sequential setup allowed the automatized clustering of the tumors into phenotypic regions based on the coregistered ADC and [18F]FDG values in every voxel, providing phenotypic maps of the tumors (Figure 4A).
Figure 4.
Multiparametric imaging approach: [18F]FDG and ADC parametric maps were calculated, and combined to phenotypic maps (A). The combination of both criteria identified 3 distinct phenotypes (A, B): solid acinar tumor regions ([18F]FDG TMR > 9.6), cystic hyperplastic regions (blue) (ADC values > 1.0 * 10−3 mm2/s) and solid nodular regions with a double-negative pattern (green; ADC < 0.6 * 10−3 mm2/s and [18F]FDG TMR < 5.1) (B). The solid nodular cluster (green) was mixed with the background population (A, B).
As expected, fusion of [18F]FDG and ADC in the same tumors enabled the identification of solid and cystic regions within the tumors (Figure 4A). Furthermore, beyond this classification, the solid regions could be differentiated into 2 distinct phenotypic patterns, solid acinar and very rare solid nodular regions. Solid acinar regions were characterized by [18F]FDG uptake values higher than 9.1 TMR; as shown previously, cystic and solid nodular regions showed low [18F]FDG uptake. Whereas the cystic areas showed high ADC values (ADC > 1.0 *10−3 mm2/s), solid regions were characterized by low ADC values. Thus, the solid nodular regions were characterized by a double-negative signature (low ADC and low [18F]FDG uptake), enabling the further differentiation of the 3 phenotypes, cystic hyperplasia, solid acinar and solid nodular malignancy, by the ADC-[18F]FDG combination (Figure 4B, Supplemental Table II).
The identification of the solid nodular phenotype was an important finding and an example of the benefits of combined PET/MRI, as this nodular malignancy was not identifiable with single-modality imaging. In the MRI it was distinguished from the background and identified as a solid region, but could not be differentiated from the acinar region, while [18F]FDG could not detect the lesion at all. Thus, if a comparable masked phenotype is present in human disease, the differentiation of this phenotype from metabolic active solid phenotypes could only be provided by the combined imaging procedure of PET/MRI.
Clinical translation: PET/MRI decodes intratumoral heterogeneity in human breast cancer
a) [18F]FDG-PET is a feasible tumor marker
To confirm the translational potential of the described approach we investigated 5 patients with biopsy-proven IBC-NST, employing simultaneous PET/MRI 7 ± 3 days before surgery. After surgical excision, pathological analysis of the whole lesions was performed.
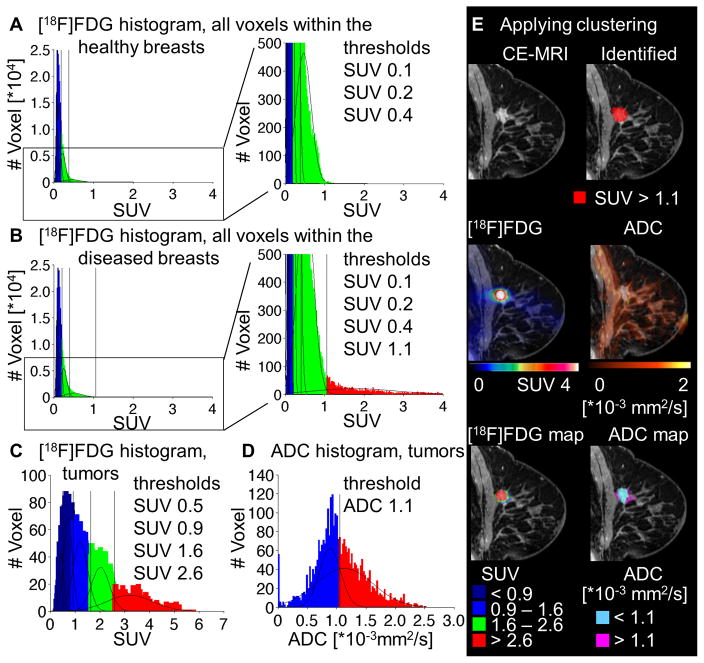
The first approach was the metabolic identification of tumor tissue within the breasts of the patients, to verify the potential of the method to identify malignant breast tissue, as described for the preclinical model. For this purpose, VOIs covering both the entire healthy and the entire diseased breasts were drawn on the basis of the CE-MRI. The clustering approach for [18F]FDG was applied separately on the sum of all voxels within the healthy (Figure 5A) and within the diseased breasts (Figure 5B). The best fit for the healthy breasts was identified for a sum of 4 Gaussian distributions, whereas the diseased breasts were best fitted by a number of 5 Gaussian distributions. The highest uptake population within the diseased breasts, including SUV > 1.1, was completely absent in the healthy breasts (Figure 5A,B).
Figure 5.
Voxelwise analysis approach for clinical [18F]FDG-PET and DW-MRI: voxel data of the healthy (A) and diseased (B) breast were analyzed by the GMM approach for [18F]FDG. The best fits were identified for a sum of 4 (A) and 5 (B) Gaussian distributions, respectively. For intratumoral clustering, voxels within all tumors were fitted for [18F]FDG with a best fit of 5 Gaussian distributions (C) and ADC with a best fit of 2 Gaussian distributions (D). A representative tumor (E): CE-MRI as well as the parametric color maps produced for the whole breast approach for [18F]FDG, applying the threshold of 1.1 in SUV (first row); ADC and PET data (second row) and the parametric maps for the intratumoral clustering for [18F]FDG and ADC (third row).
Applying this threshold of SUV > 1.1 to the whole breasts, the tumors within the breasts of all 5 patients were identified and confirmed as malignant tumors by histological analysis (Figure 5E, first row). The dimensions of the tumors given by this approach (maximal diameter, exemplary tumor: 16 × 15 × 15 mm3, Figure 5E) showed a good correlation of the identified tumors as compared to the dimensions described by post-surgical pathological analyses (maximal diameter, exemplary tumor: 16 × 13 × 10 mm3, Figure 5E, Supplemental Table III). This first approach could provide a user independent method to non-invasively identify malignant tumors.
b) Combined [18F]FDG-ADC-PET/MRI is able to cluster heterogeneous structures
As the main purpose of the study was the intratumoral classification of the lesions, we isolated all voxels within the morphologically identified tumor regions, to employ the intratumoral clustering on the sum of all tumor voxels within the 5 primary tumors. For [18F]FDG, the clustering approach identified 5 Gaussian distributions, separated by the following thresholds: SUV = 0.5, 0.9, 1.6 and 2.6 (Figure 5C). The ADC data was clustered by only 2 Gaussian distributions as the best fit, separated by the threshold of 1.1 *10−3 mm2/s, distinguishing a low diffusion population from a high diffusion population (Figure 5D). However, in this case the populations were not clearly separated, yielding a high amount of a mixed population within the blue range with low ADC values.
The thresholds were plotted back into the MRI data to illustrate the clustered populations spatially within the VOIs (Figure 5E, third row). In accordance to the preclinical data, the clinical data showed the complementary pattern of [18F]FDG uptake and ADC. Whereas the [18F]FDG uptake was higher in solid tumor regions, the ADC values were low in the solid tumor regions. However, higher ADC values were observed in the healthy gland structure of the breast and in single tumor regions, which were therefore suggested to be less solid.
As the previous approach already showed that [18F]FDG uptake values up to SUV of 1.1 represent the background uptake of healthy breast tissue, the first two populations (SUV < 0.5 and SUV 0.5 – 0.9) could not be distinguished from the background of the healthy tissue. As these populations were only present in the periphery of the tumors, surrounding the actual tumor areas, detected by the CE-MRI, they could be neglected as background populations (Figure 5E, third row).
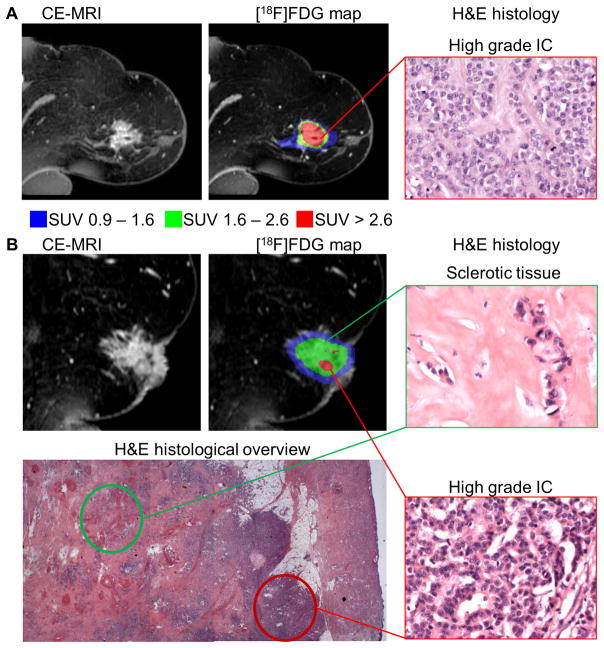
The remaining 3 uptake populations, especially the two highest populations, were considered as real tumor populations and were spatially correlated to the histological findings (Figure 6).
Figure 6.
[18F]FDG uptake populations revealed histological differences in heterogeneous tumors in the clinical setting: in a representative tumor the highest [18F]FDG accumulation represented (red, SUV > 2.6) correlated with highly aggressive IC regions in H&E histology (400 × magnification) (A). Another tumor showed large areas of lower [18F]FDG uptake populations (green, SUV 1.6–2.6 or blue, SUV 0.9 – 1.6) correlating to areas of sclerosis in the histological overview and in the green histology box (400 × magnification); smaller IC regions within this tumor in the red histology box (400 × magnification) correlated to small spots of the red population in the imaging data (B).
Comparing different regions of aggressiveness within the tumors scored by their proliferative activity and invasive growth pattern by histology, highly aggressive IC regions with high mitotic activity showed the highest [18F]FDG accumulation represented by the highest uptake population which was marked in red in the figures (SUV > 2.6).
Here, we present the tumors of 3 representative patients. Figure 6A shows a tumor consisting homogeneously of an IC, according to histology, which is mainly represented by the highest red [18F]FDG population in the PET data. On the other hand less aggressive regions with lower mitotic activity showed lower [18F]FDG accumulation, represented by the green (SUV 1.6–2.6) or blue (SUV 0.9–1.6) uptake populations. The tumor in Figure 6B contained large areas of sclerosis with less tumor cells and less mitotic activity that correlated with the green uptake population. In contrast sporadic small IC regions of high proliferation and aggressiveness within this tumor were represented by small spots of the red [18F]FDG population in the imaging data (Figure 6B). Focusing only on the CE-MRI scans, both tumors in Figure 6A and B had very similar morphology. The inter- and intratumoral biological and histological differences of these 2 tumors could only be distinguished by the metabolic PET clustering.
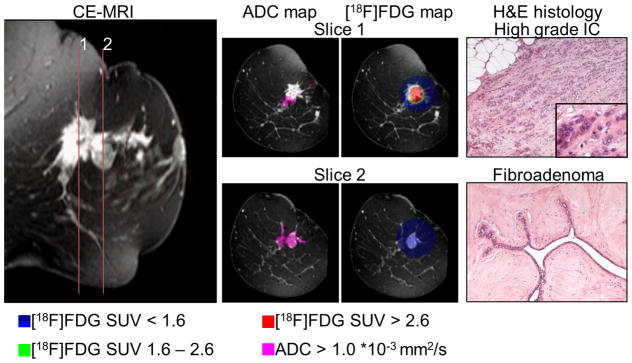
Moreover, combining [18F]FDG-PET, CE- and DW-MRI, several regions in a tumor with a very heterogeneous morphological and histological pattern, composed of multiple foci of IC as well as a fibroadenoma component, were distinguished (Figure 7). While the extent of the tumor tissue was delineated by the CE-MRI, the aggressive spots of the IC were identified as described above by the high [18F]FDG uptake population in red (SUV > 2.6), that also correlated with low ADC values (Figure 7, Slice 1). In contrast, the fibroadenoma could be distinguished from the rest of the tumor by a lower [18F]FDG accumulation (blue population, SUV 0.9–1.6) combined with the high ADC population (ADC > 1.1 *10−3 mm2/s; Figure 7, Slice 2).
Figure 7.
Combined [18F]FDG-PET and DW-MRI could distinguish several regions within heterogeneous disease: a representative tumor, composed of multiple foci of IC and a fibroadenoma component. In slice 1 the aggressive spots of the IC was identified by the high [18F]FDG population (red, SUV > 2.6). The fibroadenoma in slice 2 could be distinguished from the rest of the tumor by a low [18F]FDG accumulation (blue, SUV 0.9 – 1.6) combined with high ADC values (ADC > 1.1 *10−3 mm2/s); histological slices are shown in 100 × magnification, the insert in 400 × magnification.
This clinical data demonstrated that a comprehensive tumor profiling was possible employing combined PET/MRI in the clinical setting. Thus, the method showed great potential for the clinical translation in larger scale studies.
Discussion
Current trends in patient care aim for individualized medicine, making diagnostics and treatment strategies more complex. An important prerequisite for individualized therapies is a sophisticated diagnostic strategy that reveals even small alterations in the disease profile.
The standard diagnostic procedure in breast cancer involves mammography, sonography as well as histological validation. However, there are limitations to these techniques, for example in young patients with dense breast tissue (26). The value of PET and MRI for breast cancer diagnosis remains controversial. As recommended by the national comprehensive cancer network (NCCN) in high risk patients, MRI has clear advantages (27–30), but an improvement of patient outcome was not yet confirmed (28,31). While not considered in the NCCN guideline to date there is growing evidence for the value of PET and positron emission mammography (PEM) (32–36). Avril et al. correlated [18F]FDG uptake with several prognostic markers such as histological tumor type (ductal vs. lobular), microscopic tumor pattern (nodular vs. diffuse) and tumor proliferation (37). Yet, standard [18F]FDG-PET appears insufficient as single-information for breast cancer diagnosis (38). Other promising tracers, e.g. addressing hormone-receptor status or gastrin-releasing peptide receptor are currently under clinical investigation (32,39–41). Major restrictions in most clinical studies compared with this study are the exclusive use of single modality imaging and conventional image analysis. For example when mammography is difficult, as in high risk patients with mammographic dense breast tissue (42), the excellent soft tissue contrast of the MRI and the high sensitivity of the PET and functional MRI can provide essential information in the diagnostic process (26,37,43). Here, we showed that in combination with advanced analysis approaches, molecular and functional imaging can provide a more detailed characterization of the tumors inter- and intratumorally.
The assessment of tumor heterogeneity represents an emerging challenge in cancer diagnostics and treatment as e.g. polygenic drug resistance is a major issue in cancer patient care to date (44). Potts et al. summarized outstanding work identifying breast cancer heterogeneity and predicting therapy response and resistance on the basis of immunohistochemistry (4). To overcome the snapshot character of these biopsy-based diagnoses, in vivo diagnostic tools like MRI, CT and PET, combined with advanced analysis approaches, such as multispectral analysis or texture analysis which are described as a powerful prognostic feature to improve diagnostics, tumor staging as well as therapy response assessment in many types of cancer (45–47) must be correlated to histology. Especially MRI, including functional and spectroscopic techniques showed great potential in imaging intratumoral heterogeneity and identifying biological subpopulations within single tumors, distinguishing for example viable tumor from necrosis and adipose healthy tissues (48), or classifying the tumor microenvironment in terms of vascular heterogeneity, hypoxia and acidity (49,50).
In our preclinical study, only combined PET/MRI and advanced image analysis enabled the detection and discrimination between 2 malignant tumor phenotypes (solid nodular and acinar) from premalignant cystic hyperplasia. Especially the identified solid nodular tumor regions with very low [18F]FDG uptake would be misinterpreted as non-malignant using a single PET approach, if detected at all.
In the translational study with 5 IBC-NST tumor patients the simultaneous PET/MRI approach including the GMM analysis led to the differentiation of highly aggressive and mitotic active tumor regions to less aggressive and sclerotic regions as well as to benign lesions such as a fibroadenoma. These results pointed out the opportunities of in vivo classification of tumors in the diagnostic workflow.
Using this analysis, phenotypic identification is based on data-derived thresholds, yielding a biology-driven distinction rather than user-dependent classification, showing prospective potential, where stable thresholds derived from the training data set were successfully transferred to other tumors of the same origin in the preclinical setting. Once defined, identified and verified, the thresholds may be applied to other tumors within the same disease without the need of the complete image analysis procedure for every individual dataset. Its applicability and diagnostic value in other preclinical models as well as a larger scale validation in the clinical setting, however, was beyond the scope of this work. This analysis describes that the co-existence of different phenotypes within the same tumor can be visualized by multiparametric [18F]FDG PET/MR imaging, in both in animals as well as in patients suffering from breast cancer. This new approach has to be tested in further prospective studies; we distinguished a clinical significance to our approach.
Because treatment stratification is based on tumor biology received by minimal invasive biopsy, an accurate tissue sampling, especially in multifocal and multicentric breast tumors, is highly desirable. Therefore, our non-invasive tumor classification might enable a precise target selection and biopsy guidance in breast cancer, by revealing the presence of tumor heterogeneity and detecting the most aggressive part of the tumor, and thus would reduce the risk of under-grading and under-treatment.
Another application of this multiparametric imaging might be the early response evaluation after one to two cycles of neo-adjuvant chemotherapy (NACT), in order to adapt cancer treatment regimen or to spare ineffective and toxicity related therapy. Because a complete remission after NACT is related with longer survival, an accurate prediction of a pathologic complete response after NACT and prior to surgery is highly desirable and might have a significant impact on further treatment stratification e.g. mastectomy vs breast conserving therapy especially in multifocal/multicentric breast cancer. Finally, in cases of larger resection volumes our approach might facilitate the final histopathological assessment and enhance the diagnostic accuracy, by defining point of interest with suspicious vital/avital tumor areas that would be processed separately after the surgical procedure.
Similar combined [18F]FDG/ADC approaches showed high diagnostic value, for example, in lung and breast cancer patients (51,52) but could not provide prognostic thresholds due to the exclusive use of single value analysis in the breast cancer study and high grades of heterogeneity in the investigated lung disease, although analyzed by a GMM approach.
Current trends in cancer research showed that radiomics produce promising prognostic markers when applied on large patient cohorts (16,17,53). Our work further demonstrates that combined PET/MRI may complement these approaches by adding another biology-derived feature into the workflow, potentially moving one step further towards personalized medicine and individualized therapy planning.
Supplementary Material
Acknowledgments
Financial Support: Parts of this work were funded by fortüne grant 2223-0-0, the Swiss Werner Siemens Foundation, the European Research Council grant 323196 (ImageLink) and by Grants U01 CA141582 and U01 CA141541 from the National Cancer Institute’s Mouse Models of Human Cancers Consortium.
The authors would like to thank Maren Harant, Funda Cay, Daniel Bukala, Sandro Aidone, Carsten Groeper, Gerd Zeger, Holger Schmidt, Kerstin Fuchs, Laura Kübler and Judith E. Walls for excellent technical assistance and the radiopharmacy team for the extensive tracer syntheses. Further, the authors thank Siemens Healthcare, Erlangen, Germany for providing the dedicated 4-channel breast MRI coil for the clinical measurements.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed by the authors.
References
- 1.Tavassoli FA, Devilee P. Pathology and genetics of tumours of the breast and female genital organs. World Health Organization; 2003. [Google Scholar]
- 2.Weigelt B, Horlings H, Kreike B, Hayes M, Hauptmann M, Wessels L, et al. Refinement of breast cancer classification by molecular characterization of histological special types. The Journal of pathology. 2008;216(2):141–50. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England Journal of Medicine. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potts SJ, Krueger JS, Landis ND, Eberhard DA, Young GD, Schmechel SC, et al. Evaluating tumor heterogeneity in immunohistochemistry-stained breast cancer tissue. Laboratory Investigation. 2012;92(9):1342–57. doi: 10.1038/labinvest.2012.91. [DOI] [PubMed] [Google Scholar]
- 5.Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. The Journal of Clinical Investigation. 2011;121(10):3810. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer research. 2012;72(19):4875–82. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nature medicine. 2016;22(1):105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinn H-P, Kreipe H. A Brief Overview of the WHO Classification of Breast Tumors. Breast Care. 2013;8(2):149. doi: 10.1159/000350774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ignatiadis M, Sotiriou C. Understanding the molecular basis of histologic grade. Pathobiology. 2008;75(2):104–11. doi: 10.1159/000123848. [DOI] [PubMed] [Google Scholar]
- 10.Heusch P, Buchbender C, Köhler J, Nensa F, Gauler T, Gomez B, et al. Thoracic Staging in Lung Cancer: Prospective Comparison of 18F-FDG PET/MR Imaging and 18F-FDG PET/CT. Journal of Nuclear Medicine. 2014;55(3):373–78. doi: 10.2967/jnumed.113.129825. [DOI] [PubMed] [Google Scholar]
- 11.Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2. 0. European journal of nuclear medicine and molecular imaging. 2015;42(2):328–54. doi: 10.1007/s00259-014-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins CD. PET in lymphoma. Cancer Imaging. 2006;6(SpecA):S63. doi: 10.1102/1470-7330.2006.9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krak NC, van der Hoeven JJ, Hoekstra OS, Twisk JW, van der Wall E, Lammertsma AA. Measuring [18F] FDG uptake in breast cancer during chemotherapy: comparison of analytical methods. European journal of nuclear medicine and molecular imaging. 2003;30(5):674–81. doi: 10.1007/s00259-003-1127-z. [DOI] [PubMed] [Google Scholar]
- 14.Larson SM, Schwartz LH. 18F-FDG PET as a candidate for “qualified biomarker”: functional assessment of treatment response in oncology. Journal of Nuclear Medicine. 2006;47(6):901–03. [PubMed] [Google Scholar]
- 15.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. Journal of Nuclear Medicine. 2009;50(Suppl 1):122S–50S. doi: 10.2967/jnumed.108.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48(4):441–46. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2015:151169. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paran Y, Bendel P, Margalit R, Degani H. Water diffusion in the different microenvironments of breast cancer. NMR in Biomedicine. 2004;17(4):170–80. doi: 10.1002/nbm.882. [DOI] [PubMed] [Google Scholar]
- 19.Han S, Ackerstaff E, Stoyanova R, Carlin S, Huang W, Koutcher J, et al. Gaussian mixture model-based classification of dynamic contrast enhanced MRI data for identifying diverse tumor microenvironments: preliminary results. NMR in Biomedicine. 2013;26(5):519–32. doi: 10.1002/nbm.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy C, Cardiff R, Muller W. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and cellular biology. 1992;12(3):954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maglione JE, McGoldrick ET, Young LJT, Namba R, Gregg JP, Liu L, et al. Polyomavirus middle T–induced mammary intraepithelial neoplasia outgrowths: Single origin, divergent evolution, and multiple outcomes. Molecular Cancer Therapeutics. 2004;3(8):941. [PubMed] [Google Scholar]
- 22.Cardiff RD, Borowsky AD. Precancer: Sequentially acquired or predetermined? Toxicologic pathology. 2010;38(1):171–79. doi: 10.1177/0192623309356447. [DOI] [PubMed] [Google Scholar]
- 23.Delso G, Fürst S, Jakoby B, Ladebeck R, Ganter C, Nekolla SG, et al. Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. Journal of nuclear medicine. 2011;52(12):1914–22. doi: 10.2967/jnumed.111.092726. [DOI] [PubMed] [Google Scholar]
- 24.McLachlan G, Peel D. Finite mixture models. John Wiley & Sons; 2004. [Google Scholar]
- 25.Kemp BJ, Hruska CB, McFarland AR, Lenox MW, Lowe VJ. NEMA NU 2-2007 performance measurements of the Siemens Inveon preclinical small animal PET system. Physics in medicine and biology. 2009;54(8):2359. doi: 10.1088/0031-9155/54/8/007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sardanelli F, Giuseppetti GM, Panizza P, Bazzocchi M, Fausto A, Simonetti G, et al. Sensitivity of MRI versus mammography for detecting foci of multifocal, multicentric breast cancer in fatty and dense breasts using the whole-breast pathologic examination as a gold standard. American Journal of Roentgenology. 2004;183(4):1149–57. doi: 10.2214/ajr.183.4.1831149. [DOI] [PubMed] [Google Scholar]
- 27.NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Breast Cancer Screening and Diagnosis Version I. 2014;2014 doi: 10.6004/jnccn.2018.0083. [DOI] [PubMed] [Google Scholar]
- 28.Pilewskie M, King TA. Magnetic resonance imaging in patients with newly diagnosed breast cancer: A review of the literature. Cancer. 2014;120(14):2080–89. doi: 10.1002/cncr.28700. [DOI] [PubMed] [Google Scholar]
- 29.Kim JY, Cho N, Koo HR, Yi A, Kim WH, Lee SH, et al. Unilateral breast cancer: screening of contralateral breast by using preoperative MR imaging reduces incidence of metachronous cancer. Radiology. 2013;267(1):57–66. doi: 10.1148/radiol.12120629. [DOI] [PubMed] [Google Scholar]
- 30.Obdeijn I-M, Tilanus-Linthorst MM, Spronk S, van Deurzen CH, Monye Cd, Hunink MM, et al. Preoperative breast MRI can reduce the rate of tumor-positive resection margins and reoperations in patients undergoing breast-conserving surgery. American Journal of Roentgenology. 2013;200(2):304–10. doi: 10.2214/AJR.12.9185. [DOI] [PubMed] [Google Scholar]
- 31.Sung JS, Li J, Da Costa G, Patil S, Van Zee KJ, Dershaw DD, et al. Preoperative breast MRI for early-stage breast cancer: effect on surgical and long-term outcomes. AJR American journal of roentgenology. 2014;202(6):1376–82. doi: 10.2214/AJR.13.11355. [DOI] [PubMed] [Google Scholar]
- 32.Buck AK, Schirrmeister H, Mattfeldt T, Reske SN. Biological characterisation of breast cancer by means of PET. European journal of nuclear medicine and molecular imaging. 2004;31(1):S80–S87. doi: 10.1007/s00259-004-1529-6. [DOI] [PubMed] [Google Scholar]
- 33.Jansson T, Westlin J, Ahlström H, Lilja A, Långström B, Bergh J. Positron emission tomography studies in patients with locally advanced and/or metastatic breast cancer: a method for early therapy evaluation? Journal of clinical oncology. 1995;13(6):1470–77. doi: 10.1200/JCO.1995.13.6.1470. [DOI] [PubMed] [Google Scholar]
- 34.Humbert O, Cochet A, Coudert B, Berriolo-Riedinger A, Kanoun S, Brunotte F, et al. Role of positron emission tomography for the monitoring of response to therapy in breast cancer. The oncologist. 2015;20(2):94–104. doi: 10.1634/theoncologist.2014-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Tasaki Y, Kuwada Y, Ozawa Y, Inoue T. A preliminary report of breast cancer screening by positron emission mammography. Annals of nuclear medicine. 2015:1–8. doi: 10.1007/s12149-015-1040-0. [DOI] [PubMed] [Google Scholar]
- 36.Smyczek-Gargya B, Fersis N, Dittmann H, Vogel U, Reischl G, Machulla H-J, et al. PET with [18F] fluorothymidine for imaging of primary breast cancer: a pilot study. European journal of nuclear medicine and molecular imaging. 2004;31(5):720–24. doi: 10.1007/s00259-004-1462-8. [DOI] [PubMed] [Google Scholar]
- 37.Avril N, Menzel M, Dose J, Schelling M, Weber W, Jänicke F, et al. Glucose metabolism of breast cancer assessed by 18F-FDG PET: histologic and immunohistochemical tissue analysis. Journal of Nuclear Medicine. 2001;42(1):9–16. [PubMed] [Google Scholar]
- 38.Ambrosini V, Fani M, Fanti S, Forrer F, Maecke HR. Radiopeptide imaging and therapy in Europe. Journal of Nuclear Medicine. 2011;52(Supplement 2):42S–55S. doi: 10.2967/jnumed.110.085753. [DOI] [PubMed] [Google Scholar]
- 39.Prignon A, Nataf V, Provost C, Cagnolini A, Montravers F, Gruaz-Guyon A, et al. 68 Ga-AMBA and 18 F-FDG for preclinical PET imaging of breast cancer: effect of tamoxifen treatment on tracer uptake by tumor. Nuclear medicine and biology. 2015;42(2):92–98. doi: 10.1016/j.nucmedbio.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Mortimer JE, Bading JR, Colcher DM, Conti PS, Frankel PH, Carroll MI, et al. Functional Imaging of Human Epidermal Growth Factor Receptor 2–Positive Metastatic Breast Cancer Using 64Cu-DOTA-Trastuzumab PET. Journal of Nuclear Medicine. 2014;55(1):23–29. doi: 10.2967/jnumed.113.122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Kruchten M, Glaudemans AW, de Vries EF, Beets-Tan RG, Schröder CP, Dierckx RA, et al. PET imaging of estrogen receptors as a diagnostic tool for breast cancer patients presenting with a clinical dilemma. Journal of Nuclear Medicine. 2012;53(2):182–90. doi: 10.2967/jnumed.111.092734. [DOI] [PubMed] [Google Scholar]
- 42.Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, et al. Mammographic density and the risk and detection of breast cancer. New England Journal of Medicine. 2007;356(3):227–36. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 43.Siegmann KC, Krämer B, Claussen C. Current status and new developments in breast MRI. Breast Care. 2011;6(2):87. doi: 10.1159/000328273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehemtulla A. Overcoming intratumor heterogeneity of polygenic cancer drug resistance with improved biomarker integration. Neoplasia (New York, NY) 2012;14(12):1278. doi: 10.1593/neo.122096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davnall F, Yip CS, Ljungqvist G, Selmi M, Ng F, Sanghera B, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights into imaging. 2012;3(6):573–89. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soussan M, Orlhac F, Boubaya M, Zelek L, Ziol M, Eder V, et al. Relationship between tumor heterogeneity measured on FDG-PET/CT and pathological prognostic factors in invasive breast cancer. PloS one. 2014;9(4):e94017. doi: 10.1371/journal.pone.0094017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clinical Cancer Research. 2015;21(2):249–57. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carano RA, Ross AL, Ross J, Williams SP, Koeppen H, Schwall RH, et al. Quantification of tumor tissue populations by multispectral analysis. Magnetic resonance in medicine. 2004;51(3):542–51. doi: 10.1002/mrm.10731. [DOI] [PubMed] [Google Scholar]
- 49.Jackson A, O’Connor JP, Parker GJ, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clinical Cancer Research. 2007;13(12):3449–59. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 50.Gillies RJ, Raghunand N, Karczmar GS, Bhujwalla ZM. MRI of the tumor microenvironment. Journal of Magnetic Resonance Imaging. 2002;16(4):430–50. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt H, Brendle C, Schraml C, Martirosian P, Bezrukov I, Hetzel J, et al. Correlation of simultaneously acquired diffusion-weighted imaging and 2-Deoxy-[18F] fluoro-2-D-glucose positron emission tomography of pulmonary lesions in a dedicated whole-body magnetic resonance/positron emission tomography system. Investigative radiology. 2013;48(5):247–55. doi: 10.1097/RLI.0b013e31828d56a1. [DOI] [PubMed] [Google Scholar]
- 52.Karan B, Pourbagher A, Torun N. Diffusion-weighted imaging and 18F-fluorodeoxyglucose positron emission tomography/computed tomography in breast cancer: Correlation of the apparent diffusion coefficient and maximum standardized uptake values with prognostic factors. Journal of Magnetic Resonance Imaging. 2015 doi: 10.1002/jmri.25112. [DOI] [PubMed] [Google Scholar]
- 53.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. 2014:5. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.