Figure 5.
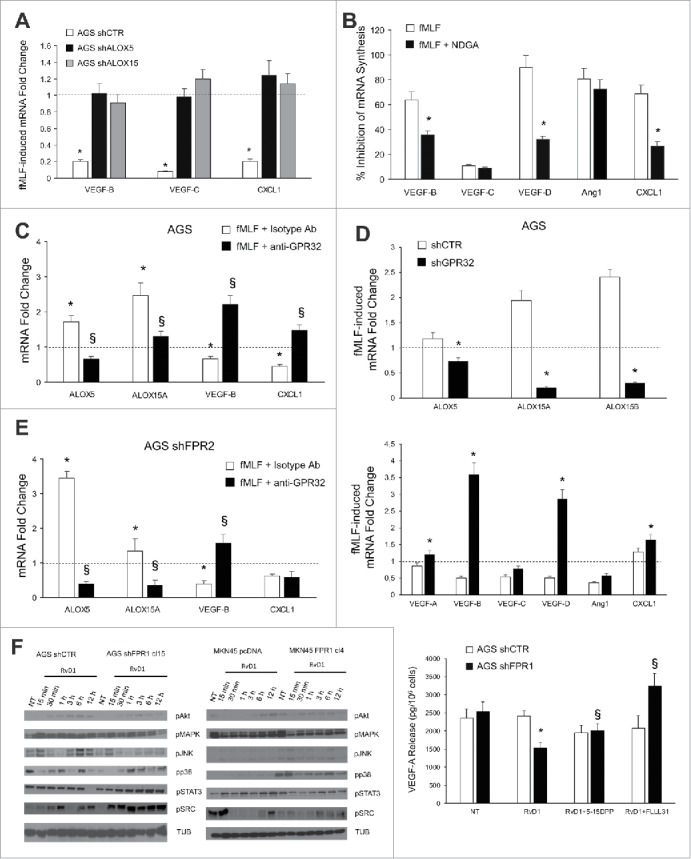
ALOXs and GPR32 are required for FPR1 tumor suppressor role. (A) VEGF-B, -C, and CXCL1 mRNA synthesis was inhibited in shCTR, but not in shALOX5 or shALOX15 AGS cells upon fMLF (10−9 M) treatment. Data are represented as mean ± SD of three independent experiments. *p < .05 vs. the relative untreated cells (dotted line). (B) Pre-treatment of AGS cells with NDGA (ALOX inhibitor) reverted the ability of fMLF to inhibit the mRNA synthesis of pro-angiogenic molecules. Data are represented as mean ± SD of three independent experiments. *p < .05 compared to fMLF treated cells. (C) In AGS cells, fMLF induced ALOX5 and ALOX15A mRNA overexpression and concomitant VEGF-B and CXCL1 mRNA downregulation. These effects were reverted by a neutralizing GPR32 antibody. An isotype-matched antibody was used as a control. Data are represented as mean ± SD of three independent experiments. *p < .05 compared to untreated cells (dotted line). §p < .05 compared to isotype-matched control. (D) fMLF significantly induced ALOX5, ALOX15A, and ALOX15B mRNA levels and significantly reduced the mRNA levels of pro-angiogenic mediators in AGS shCTR, but not in shGPR32 cells. Data are represented as mean ± SD of three independent experiments. *p < .05 compared to shCTR cells. (E) A neutralizing GPR32 antibody inhibited fMLF-induced ALOX5 and ALOX15A mRNA overexpression and concomitant VEGF-B and CXCL1 mRNA downregulation in AGS shFPR2 cells. An isotype-matched antibody was used as a control. Data are represented as mean ± SD of three independent experiments. *p < .05 compared to untreated cells (dotted line). §p < .05 compared to isotype-matched control. (F) Activation kinetics of AKT, MAPK, JNK, p38, STAT3, and SRC in AGS shCTR, AGS shFPR1, MKN45 pcDNA, MKN45 FPR1 cells assessed by western blot for their phosphorylated forms. Two STAT3 inhibitors (5-15 DPP and FLLL31) reverted the anti-angiogenic effects of RvD1 in AGS shFPR1 cells. Data are represented as mean ± SD of three independent experiments. *p < .05 compared to untreated cells (NT). §p < .05 compared to RvD1 treated cells.
