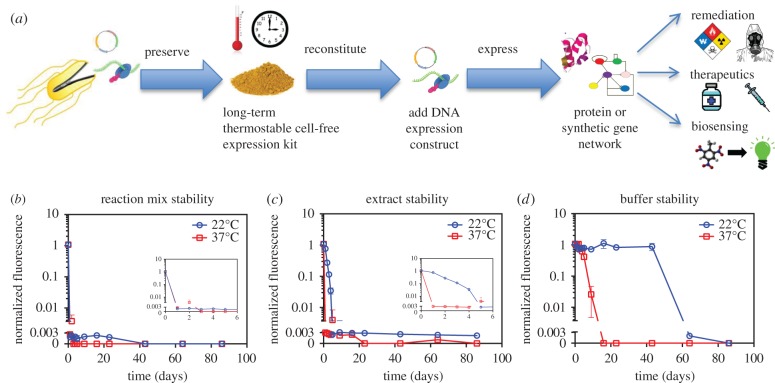
Figure 1.
Cell-free preservation overview and baseline reagent stability characterization. (a) Several applications stand to benefit from the ability to preserve cell-free protein expression reagents in a scalable, temperature stable fashion. Following reconstitution of preserved cell-free reagents, DNA encoding the expression of a protein or gene network of interest is added to enable applications in bioremediation, on demand therapeutics, and large-scale biosensing. (b) Stability characterization of cell-free reaction mixture. Aliquots of E. coli-based cell-free reaction mixtures were stored at 22°C and at 37°C. At different time points, expression capacity of the stored mixtures was assessed by adding a T7-EGFP expression construct and measuring fluorescence after 5 h of incubation. The inset zooms in on the first 6 days. (c) Stability characterization of E. coli cell extracts. Aliquots of extract were stored at 22°C and at 37°C. At different time points, fresh reaction buffer was added, and expression capacity was assessed by adding a T7-EGFP expression construct and measuring fluorescence after 5 h of incubation. The inset zooms in on the first 6 days. (d) Stability characterization of reaction buffer. Aliquots of reaction buffer were stored at 22°C and at 37°C. At different time points, fresh extract was added, and expression capacity was assessed by adding a T7-EGFP expression construct and measuring fluorescence after 5 h of incubation. (b–d) We show the means of triplicate fluorescence measurements after 5 h of incubation, with error bars indicating standard deviation. Cases where no error bars are visible indicate that error bars are smaller than the marker. Fluorescence values are normalized by dividing by the 5 h fluorescence of a standard reaction with fresh reagents. The axis break indicates the threshold for significance above background levels.