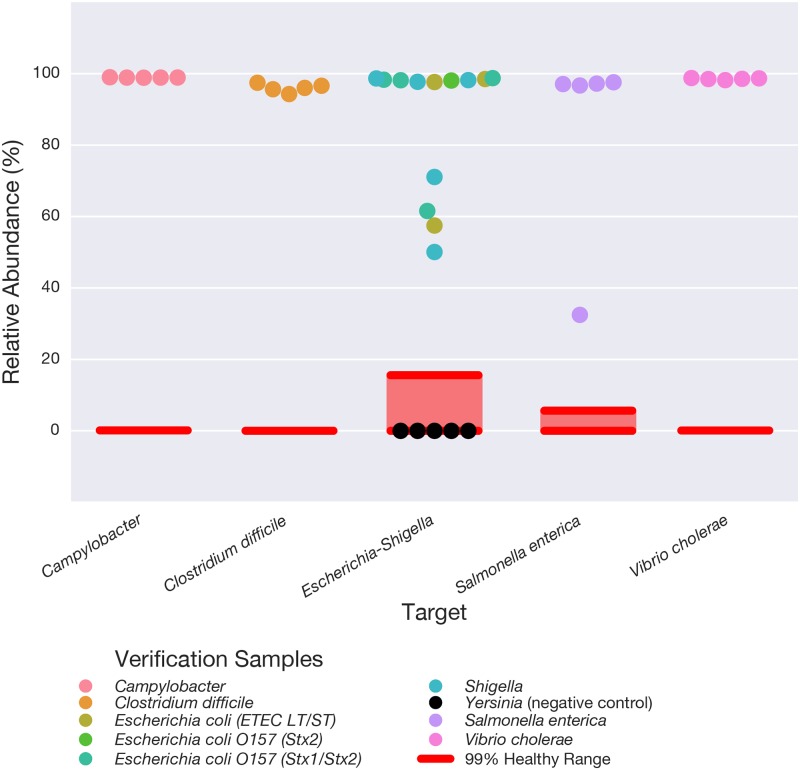
Fig 4. Experimental validation of the clinical 16S rRNA gene sequencing for pathogens on the screening test panel using verification samples.
Commercially available verification samples (Luminex) containing real or synthetic stool samples positive for at least one control taxon from the target panel were tested using the DNA extraction, amplification and bioinformatics pipeline described in this paper. Of the 35 samples on this panel, 33 yielded 10,000 or more reads. Together, these 33 samples contained the 5 pathogenic taxa in our target list, all of which were accurately identified at a level above the maximum value of the healthy range (red lines). All 33 control samples tested within the healthy range for the remainder of the taxa on our panel (not shown), and thus were considered negative for the pathogenic taxa shown here. Five samples positive for Yersinia, a genus that is not present in our target list, were included as additional negative controls. These samples are visualized for the Escherichia-Shigella genus as they contained DNA for this taxon within the healthy range.