FIGURE 1:
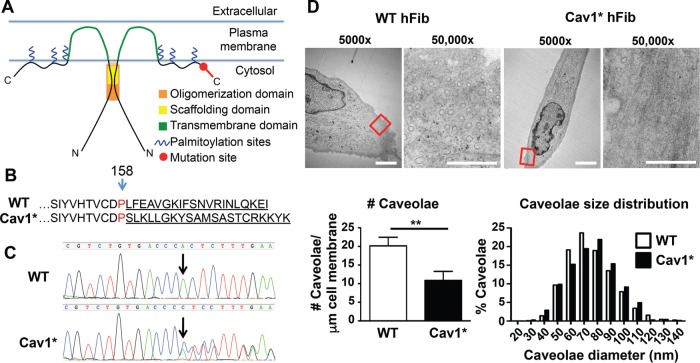
Characterization of fibroblasts carrying c.474delA mutation in caveolin-1. (A) Domains of Cav1 and location of the frameshift mutation. The oligomerization domain (residues 61–101), scaffolding domain (82–101), transmembrane domain (residues 102–134), and palmitoylation sites (133, 143, 156). (B) Amino acid sequence of Cav1*. The one-base deletion of adenine 474 causes a frameshift and changes the final 20 amino acids. (C) Sequencing of Cav1 in WT and Cav1* patient fibroblasts. Arrow indicates loss of adenine 474 due to deletion. The sequence chromatogram confirmed heterozygosity of patient cells. The one-base deletion in Cav1* sequence caused a shift that resulted in juxtaposition of both sequences after the deletion site. (D) Electron microscopy of WT and Cav1* patient fibroblasts showed lower overall number of caveolae in patient fibroblasts. Size distribution of caveolae was similar in both groups. Images were obtained from 17 cells from two different WT cell lines and 18 Cav1* cells from two different patient cell lines. For size distribution, 1140 WT and 689 Cav1* caveolae were analyzed. Scale bar, 5 μm (5000×), 0.5 μm (50,000×).