FIGURE 6:
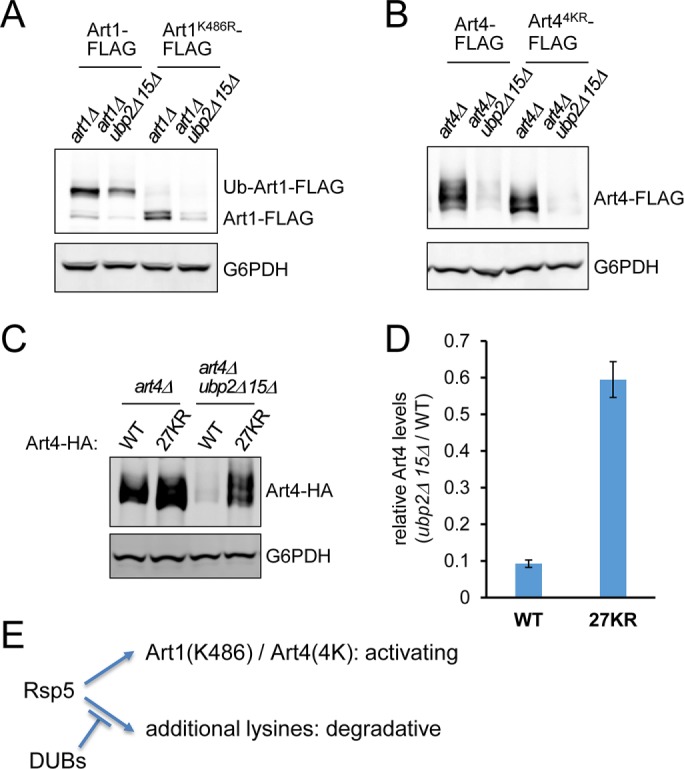
ART degradation in ubp2Δ ubp15Δ is not mediated by ubiquitination at the activating lysine residues. (A) Protein extracts of art1Δ and art1Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art1-FLAG or Art1K486R-FLAG were analyzed by SDS–PAGE and immunoblot. (B) Protein extracts of art4Δ and art4Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art4-FLAG or Art4K(235/245/264/267)R-FLAG were analyzed by SDS–PAGE and immunoblot. (C) Protein extracts of art4Δ and art4Δ ubp2Δ ubp15Δ cells expressing plasmid-encoded Art4-HA or Art427KR-HA were analyzed by SDS–PAGE and immunoblot. (D) Relative Art4 levels (art4Δ ubp2Δ ubp15Δ cells vs. art4Δ cells) as in C were quantified using the LI-COR system. Data from three independent experiments are presented as mean ± SD. (E) Model for functionally distinct ubiquitinations of ARTs. Rsp5 ubiquitinates Art1 at K486 and Art4 at K235/K245/K264/K267, which are required for their activation. In the absence of Ubp2 and Ubp15, additional lysines on ARTs are ubiquitinated, causing their degradation.
