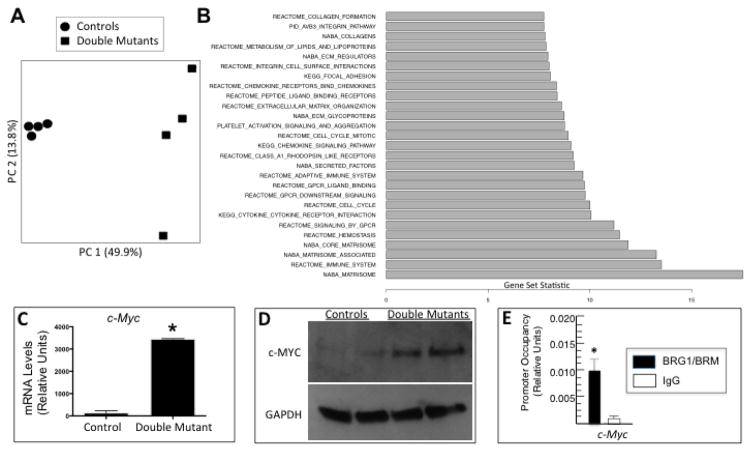
Figure 4.
BRG1/BRM transcriptional targets including c-Myc. (A) Principal component (PC) analysis of controls and Brg1/Brm double mutants based on their transcriptome profiles. (B) Significantly enriched pathways among the genes differentially expressed between Brg1/Brm double mutants and controls. All of the listed pathways have an FDR < 0.05, and the gene set statistic on the x-axis represents the z score transformation of the mean of all genes in a set. (C) RT-qPCR analysis of c-Myc mRNA levels normalized to Gapdh in control and double-mutant hearts. Data are presented as means ± SEM based on 5 independent experiments with significant differences indicated (*p<0.05). (D) Western blot analysis of heart protein lysates from controls and double mutants probed with antibodies specific for c-MYC and GAPDH as a loading control. (E) Quantitative ChIP assays demonstrating BRG1/BRM occupancy at the c-Myc promoter in wild-type mouse heart. Histograms show the relative enrichment by comparing each ChIP sample to input by qPCR (means ± SEM for three independent samples, (*p<0.05).