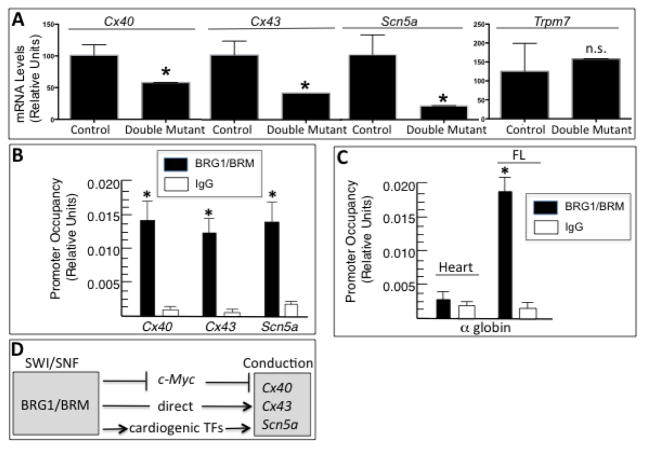
Figure 6.
BRG1/BRM regulation of conduction genes. (A) RT-qPCR analysis of mRNA levels of cardiac connexins (Cx40, Cx43) and ion channels (Scn5a, Trpm7) normalized to Gapdh in control and double-mutant hearts. Data are presented as means ± SEM based on 5 independent experiments with significant differences indicated (*p<0.05). (B, C,) Quantitative ChIP assays demonstrating BRG1/BRM occupancy at the Cx40, Cx43, and Scn5a promoters in wild-type mouse heart tissues (B) and at the α-globin locus in wild-type heart and fetal liver (FL) tissues (C). IgG immunoprecipitations serve as a negative control. Histograms show the relative enrichment by comparing each ChIP sample to input by qPCR (means ± SEM for three independent samples, (*p<0.05). (D) Working model. The BRG1 and BRM catalytic subunits of SWI/SNF complexes directly and indirectly activate the expression of Cx40, Cx43, and Scn5a to facilitate conduction in cardiomyocytes. The direct regulation is based on ChIP assays demonstrating occupancy at each promoter. The indirect regulation is mediated by inhibition of an inhibitor (c-Myc) and activation of an activator (cardiogenic transcription factors Tbx, Nkx2-5, Mef2c).