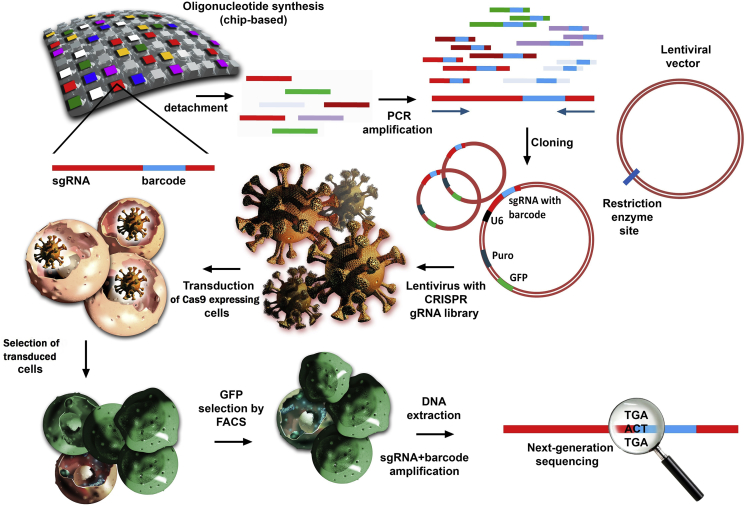
Figure 3.
Generation and Quality Control of sgRNA Libraries for CRISPR/Cas9-Mediated Phenotypic Screening
The first step illustrates the synthesis of sgRNA libraries consisting of a heterogeneous population of sequences approximately 100 bases in length with a reduced number of mutations. Quality control is significantly facilitated when a barcode of a known sequence is incorporated into each sgRNA and analyzed at the final stages through next-generation sequencing (NGS). After detachment of oligonucleotides from the solid support, the targeted sequences are amplified through PCR and then cloned in lentiviral vectors that contain a selection marker for antibiotic resistance (e.g., puromycin) and also an enrichment sequence that can be detected with the help of fluorescence-activated cell sorting (FACS) (e.g., GFP). Every sgRNA is under the activity of an U6 promoter that will facilitate its expression. The entire complex is packed in viruses that are further used for the transduction of the target cells. The first selection consists of the capacity of cells to survive in an antibiotic-enriched medium because of their integrated gene for puromycin or any other antibiotic that is used in the experiment. The second selection consists of the enrichment of the transduced cell population through GFP selection by FACS. The remaining cells that contain the viral construct are then collected for DNA extraction and analyzed through NGS, which detects individual sgRNAs because of the inserted barcode.