Figure 8.
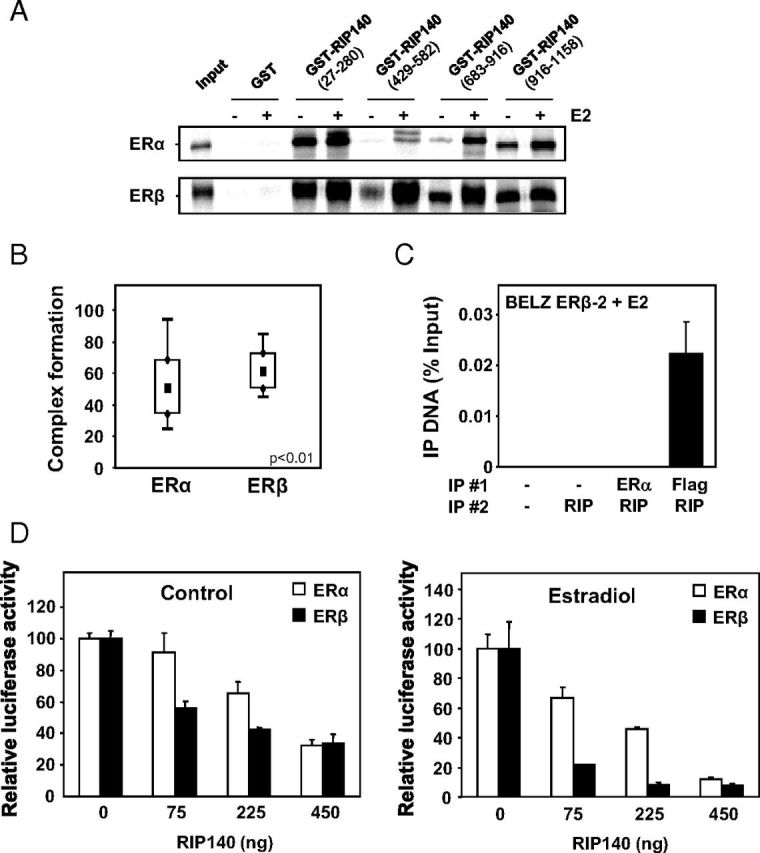
Interaction of RIP140 with ERα and ERβ. A, In vitro interaction of RIP140 with ERα and ERβ was assessed by GST pulldown assay between bacterially expressed GST-RIP proteins vs GST alone and 35S-labeled ERα or ERβ in the presence or not of E2 used at the concentration of 1 μM. B, Complex formation of RIP140 with ERα and ERβ was measured by FCCS in E2-treated COS-7 cells transfected with expression vectors for tagged-version of RIP140 and either ERα or ERβ. FCCS measurements and data analysis were conducted as described in Materials and Methods. Results are presented as percent complex formation between RIP140 and each of the ERs. C, RIP140 recruitment by the 2 ERs was performed using ChIP analysis. Two sequential immunoprecipitations (IP 1 and 2) were performed using RIP140 and Flag or ERα antibodies in BELZ ERβ-2 cell lines after 1 hour of E2 treatment (10 nM). The PCR amplification of the ERE site in front of the luciferase gene was performed as described in Materials and Methods. Values represent percentage of input. D, The effect of RIP140 on ERα and ERβ transactivation was assayed in transient transfection experiments. HeLa cells were transiently transfected with the ERE-βGlob-Luc reporter construct (500 ng) together with 500 ng of expression vector for ERα (white symbols) or ERβ (black symbols) and increasing concentrations of RIP140 expression plasmid (adjusted with empty vector). Cells were then treated with E2 for 24 hours, and luciferase activity was quantified as indicated in Materials and Methods. Results are expressed as relative luciferase activity (% of control) and are the mean (±SD) of 8 values from 3 independent experiments.
