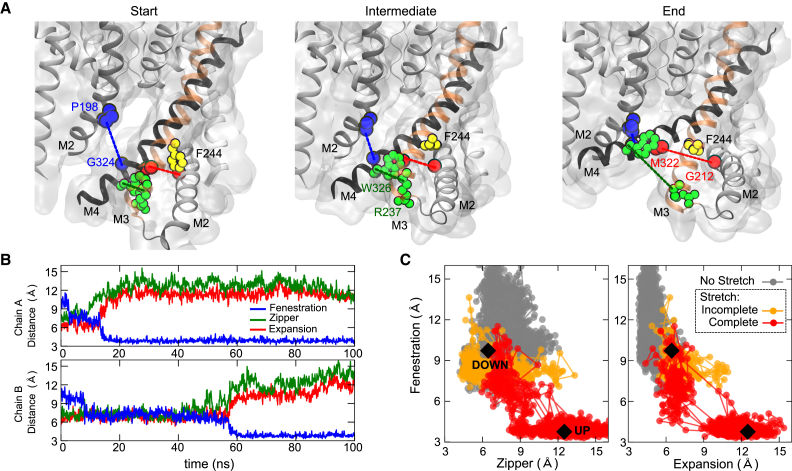
Figure 3.
Sequential Rearrangement of the TM Helices during Membrane Stretch
(A) Movement of TMs measured as a change in three distances: (1) “Fenestration” between G324 and P198 of the adjacent subunit (blue), (2) “Zipper” between W326 and R237 (green), and (3) “Expansion” between M322 and G212 (red). These changes are shown at the start, during, and end of 100 ns stretch simulation. Note unzipping of interactions between M4 and M3 requires reorientation of W326 (green) on M4; also F244 (yellow) then rotates to fill the space created by the M2-M4 expansion.
(B) Change in these distances in separate chains of TREK-2 during a stretch simulation. Note the sequence: an initial contraction of the fenestration followed by unzipping, expansion, and then full closure of the fenestration.
(C) Correlation plot comparing the change in fenestration and zipper distances during an unstretched (gray) and stretched (red) simulation sampled every 1 ns. Distances in the TREK-2 crystal structures shown as black diamonds. Within the unstretched bilayer TREK-2 samples conformations close to the down state crystal structure (PDB: 4XDJ) and does not approach the up state conformation (PDB: 4BW5). When stretched (red), the structure samples conformations similar to the up state. In some cases incomplete closure of the fenestration occurs (orange) due to obstruction by lipid tails (see also Figures 6 and S5). The right-hand panel shows a similar comparison of fenestration and expansion distances for both stretch and unstretched simulations.