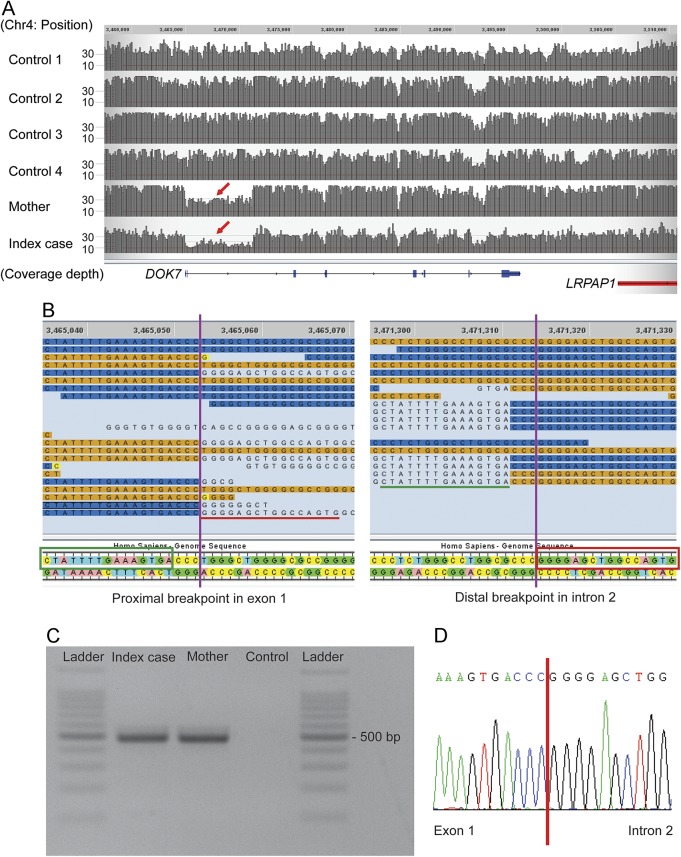
Figure 1. Whole-genome sequencing analysis and allele-specific PCR.
(A) Both index case and his mother show reduced read depth (coverage) from exon 1 to deep intron 2 of the DOK7 gene (red arrow). Controls 1–4 correspond to samples sequenced and analyzed through the same pipeline and without the diagnosis of congenital myasthenic syndromes. (B) Split reads were observed at both presumed breakpoints. Nucleotides matching the reference sequence of DOK7 are highlighted in orange/blue. Single unmatched nucleotides are highlighted in yellow, and further unmatched sequences are not highlighted. The unmatched sequence (indicated with red/green underline) of the split reads of the proximal breakpoint aligns to the reference sequence (indicated in green/red boxes) at the distal breakpoint, and vice versa. (C) The expected products amplified by allele-specific PCR were identified in the index case and the mother. (D) The junction of the breakpoint in the allele with the intragenic deletion was confirmed by Sanger sequencing of the PCR product. Coverage and reads were drawn by the graphical user interface of Sequence Miner 5.21.1 (WuXi NextCODE).