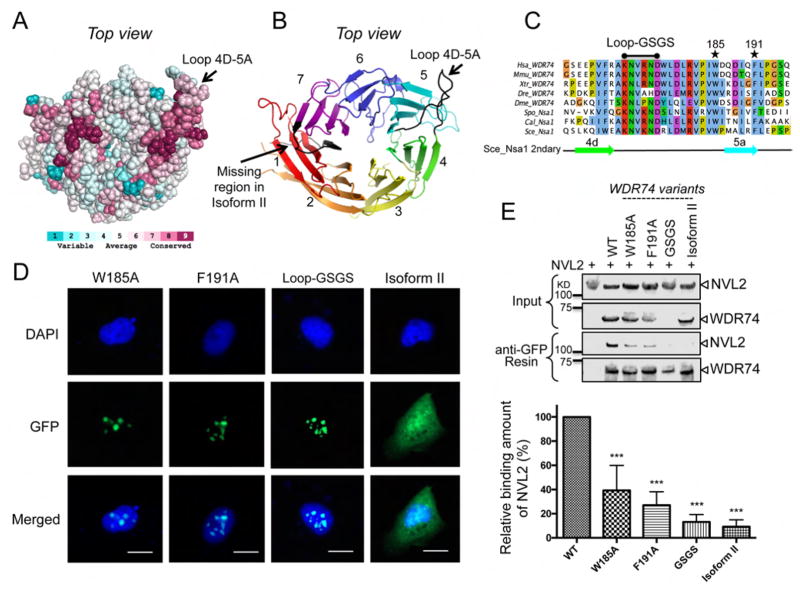
Figure 5.
Nsa1/WDR74 associates with Rix7/NVL2 (A) A surface representation of Nsa1 illustrating conserved (dark pink) to not-conserved residues (cyan). The figure was produced using the consurf website http://consurf-hssp.tau.ac.il (B) Cartoon of top view of Nsa1 highlighting the missing region of WDR74 isoform II (black) and the conserved loop (black) between blade 4D and blade 5A. (C) Selected alignment of Nsa1/WDR74 between blade 4D and blade 5A (see Figure S2 for full alignment) from different homologues of Homo sapiens (Hsa), Mus musculus (Mmu), Xenopus tropicalis (Xtr), Danio rerio (Dre) Drosophila melanogaster (Dme), Schizosaccharomyces pombe (Spo), Candida albicans (Cal), and Saccharomyces cerevisiae (Sce). The asterisk indicate the position of the point mutations in Hsa WDR74 and the black line indicates the position of the loop-GSGS (D) U2OS cells were transfected with plasmids harboring WDR74-GFP mutants and then fixed and stained with DAPI (blue). The 20 μm scale bar is representative for all panels. (E) Western blot analysis of the interaction between NVL2 with various mutations of WDR74. The relative amount of NVL2 bound to the resin is quantified below (39.3% for W185A, 27.1% for F191A, 13.2% for Loop-GSGS, and 9.2% for Isoform II) from 5 independent co-immunoprecipitations (*P < 0.05; **P < 0.01; ***P < 0.001).