FIG 2 .
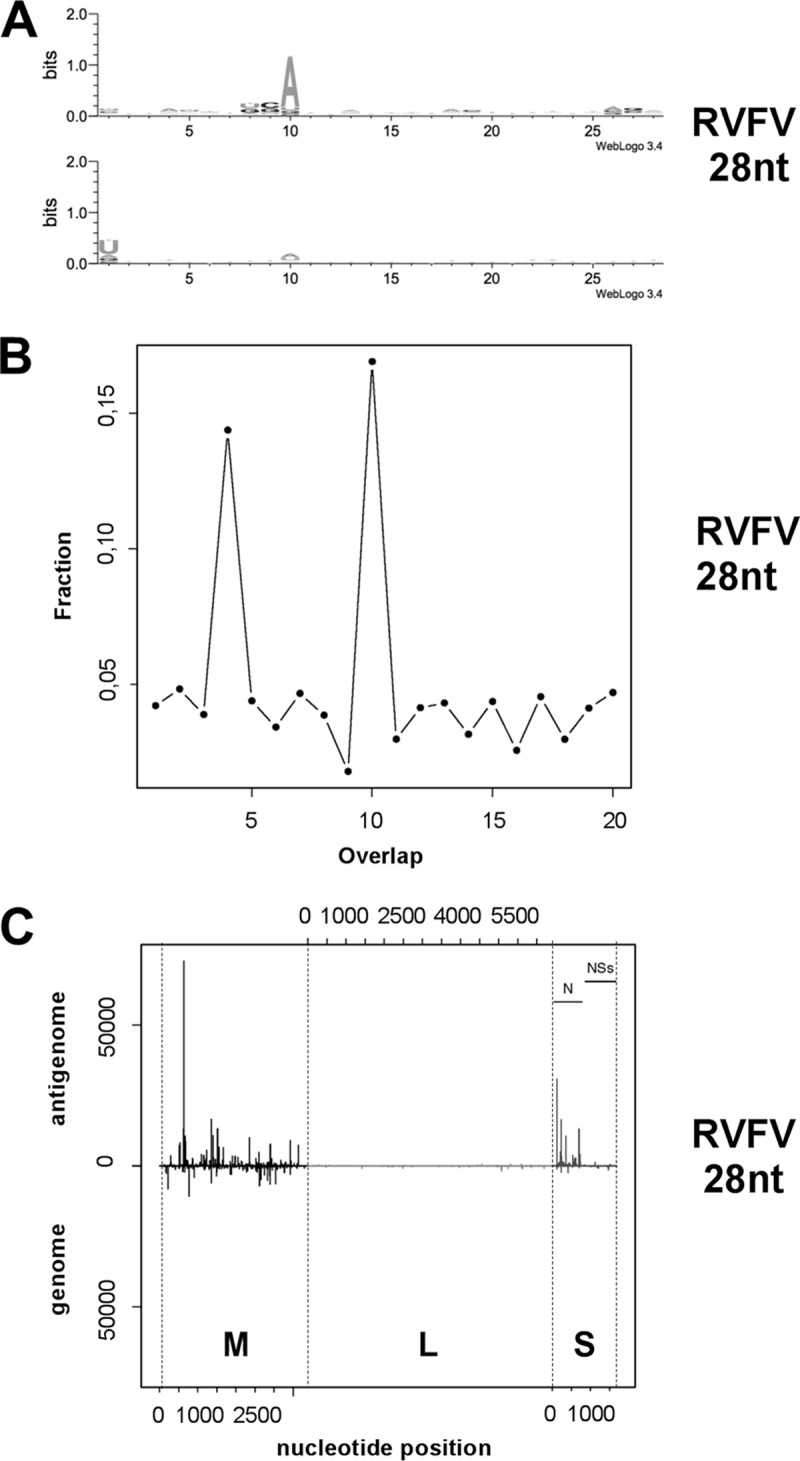
RVFV-specific piRNA-like small RNAs are produced in infected Aedes aegypti Aag2 cells. (A) Representation of piRNA sequence logos within the 28-nt reads. All reads matching the RVFV MP12 genomic and antigenomic sequences were analyzed for their base frequencies at each position. The x axis shows the nucleotide position. The y axis represents the frequency of each nucleotide at the corresponding position. (B) The panel shows the probability of overlap at the 5′ end of the opposite viral small RNA for the 28-nt reads. The x axis indicates the number of nucleotides overlapping between the antigenomic and genomic sequences; the y axis shows the mean fractions of reads overlapping. (C) Alignment of the 28-nt small-RNA reads to the RVFV MP12 sequences of the three viral RNA segments. The x axis represents the position of small-RNA reads in reference to the antigenome RNA sequence (5′-to-3′ orientation) or to the genome (3′-to-5′ orientation). The y axis represents the number of reads for each position, with reads matching the antigenome shown on the positive y axis and reads matching the genome shown on the negative y axis.
