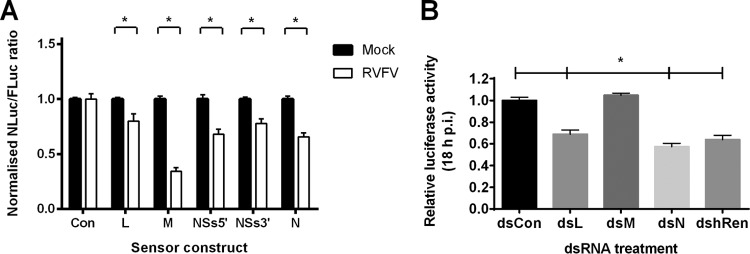
FIG 5 .
RVFV-derived small RNAs display antiviral properties. (A and B) The accessibility of RVFV mRNAs to RVFV-specific small RNAs and their antiviral properties were investigated using luciferase reporter-based sensor constructs (A) and RVFV-targeting dsRNAs (B), respectively. (A) Aag2 cells were either infected with RVFV MP12 (white bars) or left uninfected (black bars). At 24 h p.i., cells were transfected with small-RNA sensors (pIZ containing a nanoluciferase [NLuc] ORF fused to RVFV-specific/control sequences) and a transfection control (pIZ-FLuc). RVFV-specific sequences consisted of fragments amplified from the L open-reading frame (ORF; 515 nt), the M ORF (529 nt), the N ORF (498 nt), or the 5′ or 3′ halves of the NSs ORF (NSs5′, 416 nt; NSs3′, 329 nt). Primer binding sites for the respective fragments are indicated in Table S3. NLuc/FLuc ratios in transfected infected cells were normalized against the respective transfected uninfected cells. The graph presents means and standard errors of results from three independent experiments, each performed in triplicate. Statistical analysis was performed using the t test. *, P < 0.05. (B) dsRNAs targeting the RVFV rMP12delNSs:hRen L, M, and S segments (dsL, dsM, dsN, dshRen) or a control dsRNA targeting eGFP (dsCon) were transfected into Aag2 cells, followed by virus infection. Viral replication was measured by luciferase assay, and dsL, dsM, dsN, and dshRen transfected samples were normalized against the control samples. The graph presents means and standard errors of results from three independent experiments, each performed in triplicate. Statistical analysis was performed using the t test. *, P < 0.05.