Abstract
Background
Genome-wide association studies have identified the ORM (yeast)-like protein isoform 3 (ORMDL3) gene locus on human chromosome 17q to be a highly significant risk factor for childhood-onset asthma.
Objective
We sought to investigate in vivo the functional role of ORMDL3 in disease inception.
Methods
An Ormdl3-deficient mouse was generated and the role of ORMDL3 in the generation of allergic airways disease to the fungal aeroallergen Alternaria alternata was determined. An adeno-associated viral vector was also used to reconstitute ORMDL3 expression in airway epithelial cells of Ormdl3 knockout mice.
Results
Ormdl3 knockout mice were found to be protected from developing allergic airways disease and showed a marked decrease in pathophysiology, including lung function and airway eosinophilia induced by Alternaria. Alternaria is a potent inducer of cellular stress and the unfolded protein response, and ORMDL3 was found to play a critical role in driving the activating transcription factor 6–mediated arm of this response through Xbp1 and downstream activation of the endoplasmic reticulum–associated degradation pathway. In addition, ORMDL3 mediated uric acid release, another marker of cellular stress. In the knockout mice, reconstitution of Ormdl3 transcript levels specifically in the bronchial epithelium resulted in reinstatement of susceptibility to fungal allergen–induced allergic airways disease.
Conclusions
This study demonstrates that ORMDL3, an asthma susceptibility gene identified by genome-wide association studies, contributes to key pathways that promote changes in airway physiology during allergic immune responses.
Key words: ORMDL3, asthma, unfolded protein response, uric acid, Alternaria
Abbreviations used: AAD, Allergic airways disease; AAV, Adeno-associated viral vector; AHR, Airway hyperreactivity; ATF, Activating transcription factor; BALF, Bronchoalveolar lavage fluid; EGFP, Enhanced green fluorescent protein; ER, Endoplasmic reticulum; GWAS, Genome-wide association study; IRE, Inositol requiring ER-to-nucleus signal kinase; KO, Knockout; ORMDL, ORM-1 like protein; PERK, Double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase; SERCA2b, Sarco-endoplasmic reticulum Ca2+ ATPase 2b; SNP, Single nucleotide polymorphism; UK, United Kingdom; UPR, Unfolded protein response; WT, Wild-type; Xbp1, X-box binding protein 1
Graphical abstract
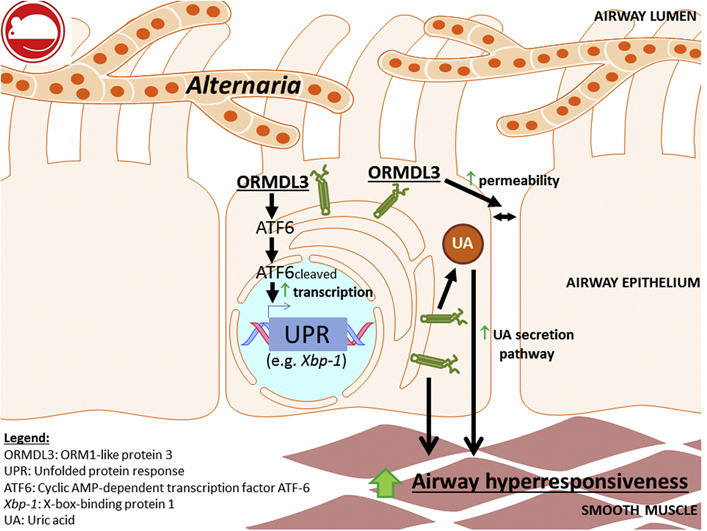
Asthma encompasses a range of pulmonary disease phenotypes commonly defined by a type 2 inflammatory response to airborne allergen concomitant with airway hyperresponsiveness (AHR).1 Although the underlying etiologies for asthma are incompletely understood, it is recognized that the epithelial barrier plays a central role in asthma pathogenesis and that genetic predisposition can be pivotal for asthma development.2 A pioneering genome-wide association study (GWAS) identified single nucleotide polymorphisms (SNPs) on chromosome 17q21 that were strongly linked to asthma.3 The same SNPs were also found to be associated with increased expression of transcripts of the ORM-1 like protein 3 (ORMDL3) gene.3 However, the functional role of ORMDL3 in asthma has not as yet been fully elucidated, and it is not known how this molecule contributes to disease pathophysiology.
ORMDL3 is expressed in various tissues, including the lung where it is predominantly localized to airway epithelial cells in response to Alternaria or ovalbumin challenge.4, 5 It is a transmembrane protein located in the endoplasmic reticulum (ER)4 where physiologically it acts to negatively regulate sphingolipid synthesis.6 Intriguingly, it has been demonstrated that reduction of sphingolipid synthesis with pharmacological compounds increases AHR in mice and human bronchial tissue.7 However, in a murine model of house dust mite–induced allergic airways disease (AAD), elevated levels of ORMDL3 in response to allergen correlate with increased ceramide production.8 Previous studies with mice universally overexpressing ORMDL3 have demonstrated that these mice spontaneously develop increased AHR and airway remodeling, which precedes elevated type 2 pulmonary inflammation.9
ER stress can be induced on perturbation of luminal Ca2+ levels within the ER, consequently decreasing the efficiency of correct protein folding.10 Accumulation of nascent or misfolded protein in the ER lumen leads to activation of the unfolded protein response (UPR).11, 12 Available data indicate that ORMDL3 modulates the UPR although the pathways involved may be tissue specific. Bone marrow–derived macrophages from Ormdl3 transgenic mice have increased activation of the activating transcription factor (ATF) 6 pathway of the UPR,9 similar to previous observations using lung epithelial A549 cells transfected with ORMDL3 in vitro.5 In these systems, expression of ORMDL3 is positively correlated with expression of sarco-endoplasmic reticulum Ca2+ ATPase 2b (SERCA2b), a downstream target of the ATF6 pathway.13 In HEK293 and Jurkat cells transfected with Ormdl3, the data illustrate that ORMDL3 binds to and inhibits SERCA, thereby impeding entry of cytosolic Ca2+ into the ER lumen and initiating the UPR.14 However, within this experimental setting, the main impact of ORMDL3 is on the double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase (PERK)/eIF2α arm of the UPR.
In the present study, Ormdl3-deficient mice were generated to investigate the role of ORMDL3 in determining the pulmonary response to allergen. In addition, we have reconstituted ORMDL3 specifically in the bronchial epithelium of these knockout (KO) mice to ascertain the specific contribution of epithelial ORMDL3. These mice reveal that ORMDL3 is pivotal in the generation of fungal allergen–induced AHR via modulation of cellular stress pathways. Our data reinforce the findings of asthma GWAS, placing ORMDL3 as an important mediator in the development of allergen-induced AHR.
Methods
Generation of Ormdl3 KO mice
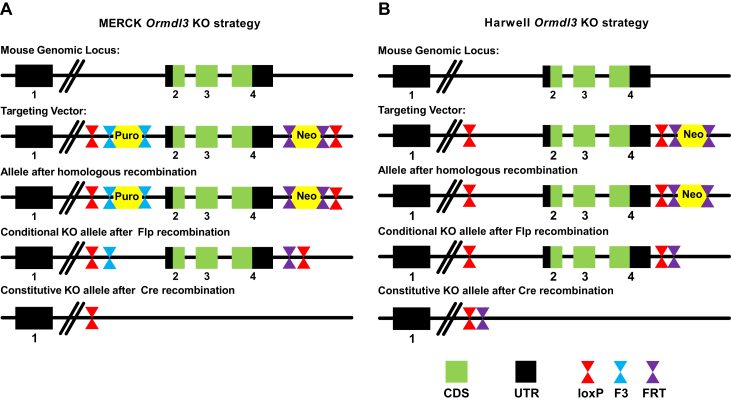
Two separate Ormdl3 alleles were used in the study. The first was generated by Dr. Y Zhang at the Mary Lyon Centre, MRC Harwell, and the second was made and kindly donated by Merck Research Laboratories. Both constructs contained LoxP sites flanking exons 1 to 4 of Ormdl3, the translation initiation site of the gene being contained within exon 2. Initially, conditional alleles were generated by Flp-mediated removal of selection cassettes. The targeting strategies for both lines were similar; however, the Merck construct contained dual Neo and Puro selection cassettes, whereas the Harwell allele contained a single Neo selection cassette (see Fig E1 in this article's Online Repository at www.jacionline.org). At Merck, targeting constructs were micro-injected into embryonic stem cells from C57/Bl6NTac mice and complete Ormdl3 KOs were subsequently generated by Cre-mediated recombination, using globally expressed Cre-driver mouse lines.15 At Harwell, targeting constructs were micro-injected into ES cells from R1 129 ES cells and Ormdl3 KOs were subsequently generated by Cre-mediated recombination, using Tg(ACTB-cre)3Mrt mice (Infrafrontier; accession no. EM:06107). These Ormdl3 KO mice were subsequently crossed on to a C57BL/6J background (10 generations). A similar response to allergen was observed in both strains. The MERCK Ormdl3 KO line (Ormdl3 KOMer) was used for experiments where mice were treated as adults. Experiments which required in-house breeding at Imperial College were performed using the Ormdl3 KO line generated by Dr Y Zhang (Ormdl3 KOHar).
Fig E1.
Comparative schematic of strategies used to generate Ormdl3MER KO and Ormdl3HAR KO mice. A, Strategy used by MERCK. The gene Ormdl3 contains 4 exons of which exons 2 to 4 are encoding the ORMDL3 protein. Exons 2 to 4 were consequently flanked by 2 antibiotic selection cassettes (Puro, puromycin; Neo, neomycin) and loxP recombination sites were inserted upstream of the Puro cassette and downstream of the Neo cassette. Following homologous recombination, Flp recombination, and Cre recombination steps, the resulting Ormdl3MER KO strain retains a loxP recombination site downstream of exon 1. B, The approach used by MRC Harwell slightly differed from the MERCK strategy, as a single neomycin selection cassette was included in the targeting vector. LoxP recombination sites were located between exons 1 and 2 and between exon 4 and the FRT-flanked Neo cassette. The resulting Ormdl3HAR KO mice strain therefore retains a loxP, as well as an FRT recombination site after homologous, Flp and Cre recombination. CDS, Coding sequence; UTR, untranslated region.
Construction of enhanced green fluorescent protein and Ormdl3-enhanced green fluorescent protein adeno-associated viral vectors
An Ormdl3-enhanced green fluorescent protein (EGFP) (N-terminal tag) gene fusion was constructed and cloned into plasmid pZac2.1 under control of the ubiquitous cytomegalovirus promoter. Adeno-associated viral vectors (AAVs) were produced by Penn Vector Core (University of Pennsylvania, Pa). On day 5 of life, neonatal mice were intranasally administered 1 × 1011 genome copies of either AAV EGFP (control vector) or AAV Ormdl3-EGFP (AAV Ormdl3) with 50 mU neuraminidase (Sigma-Aldrich, Dorset, United Kingdom [UK]) in PBS. At age 8 weeks, mice received either 10 μg Alternaria alternata extract (Greer Laboratories, Lenoir, NC) or PBS via intranasal instillation, 3 times per week for 5 weeks. Mice were killed 18 hours postfinal Alternaria instillation.
Induction of AAD
Ten- to 12-week-old male wild-type (WT) and Ormdl3 KOMer mice were bred at Taconic Biosciences (Germantown, New York, NY). WT and Ormdl3 KOHar mice (Harwell, UK) were bred at Imperial, and neonatal mice were used for overexpression studies. All mice were housed in specific pathogen-free conditions and given food and water ad libitum. Mice were exposed to 20 μg of purified Alternaria alternata extract (Greer Laboratories) (in 25μL PBS) intranasally 3 d/wk for 5 weeks. All procedures were conducted in accordance with the Animals (Scientific Procedures) Act 1986.
Assessment of airway function
AHR in response to increasing doses of methacholine (10-200 mg/mL; Sigma-Aldrich) was measured as previously described,16 using the Flexivent system (Scireq, Montreal, Quebec, Canada).
In vitro measurement of airway smooth muscle function
Tracheal tissue was harvested from WT or Ormdl3 KOMer mice. Agonist concentration-response curves to methacholine or vehicle were subsequently fitted by least-squares, nonlinear regression based on the Hill equation (Prism 5, GraphPad Software Inc, La Jolla, Calif). Mean EC50 values were calculated by averaging data from interpolation of response curves constructed for each individual tissue within a data set.
Statistical analysis
All data were analyzed using Graph Pad Prism 6 (GraphPad Software). Box and whisker plots depict the median and interquartile range and minimum and maximum values. Line graphs and bar charts are expressed as mean ± SEM, and data were analyzed using nonparametric Mann Whitney U tests with significance defined as *P < .05, **P < .01, and ***P < .001.
Methods for in vitro assessment of airway smooth muscle function, quantitative PCR, RT-PCR, immunofluorescence, isolation and sorting of epithelial and CD45+ cells, flow cytometry, Western blotting, and mediator analysis are described in the Methods section in this article's Online Repository at www.jacionline.org.
Results
Ormdl3 deficiency protects mice from developing fungal allergen–induced AHR
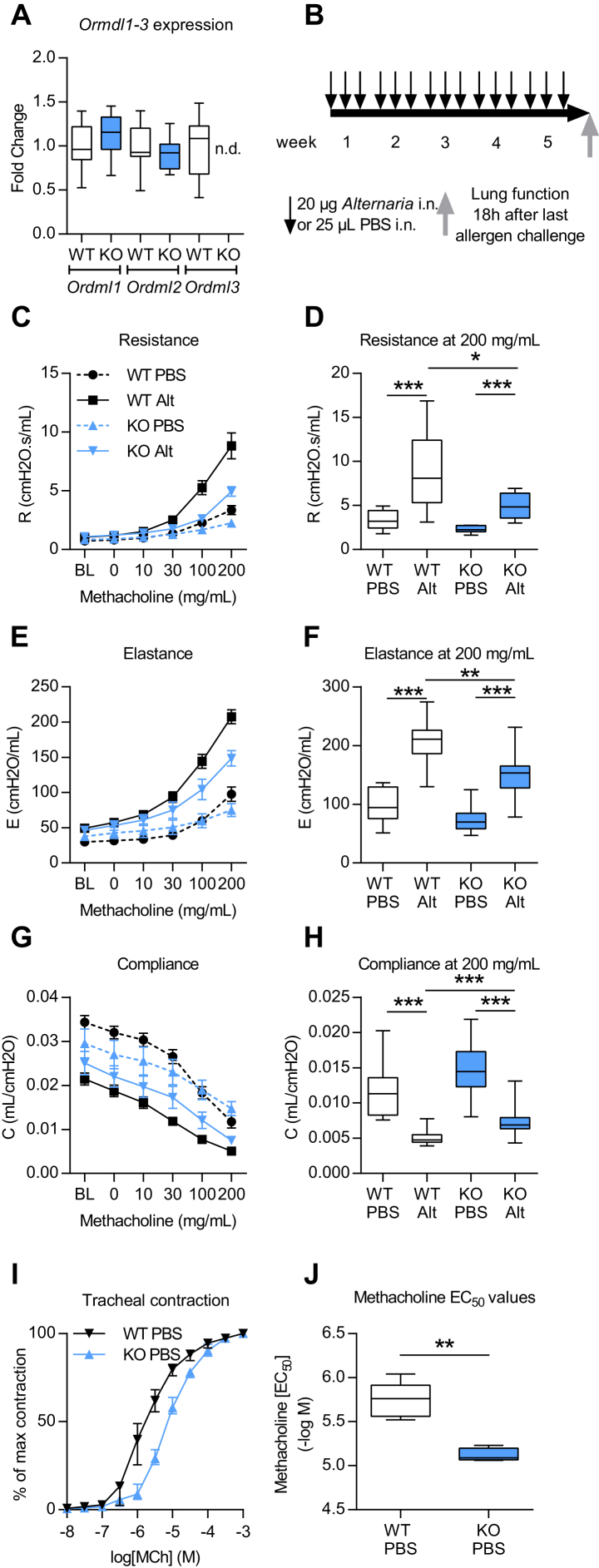
ORMDL3 is ubiquitously expressed in adult and fetal tissues including the lung3, 4 and also in leukocytes.5, 17 To assess the impact of ORMDL3 on induction of AAD, we generated global Ormdl3 KO mice. We report that mice lacking Ormdl3 had no baseline phenotype, appearing healthy and fertile with no alterations in the lung architecture or immune changes at homeostasis compared with WT littermate C57bl/6 mice. Despite deletion of Ormdl3, there was no compensatory upregulation of the closely related genes Ormdl1 and Ormdl2 in the lungs of the KO mice as determined by quantitative PCR (Fig 1, A).
Fig 1.
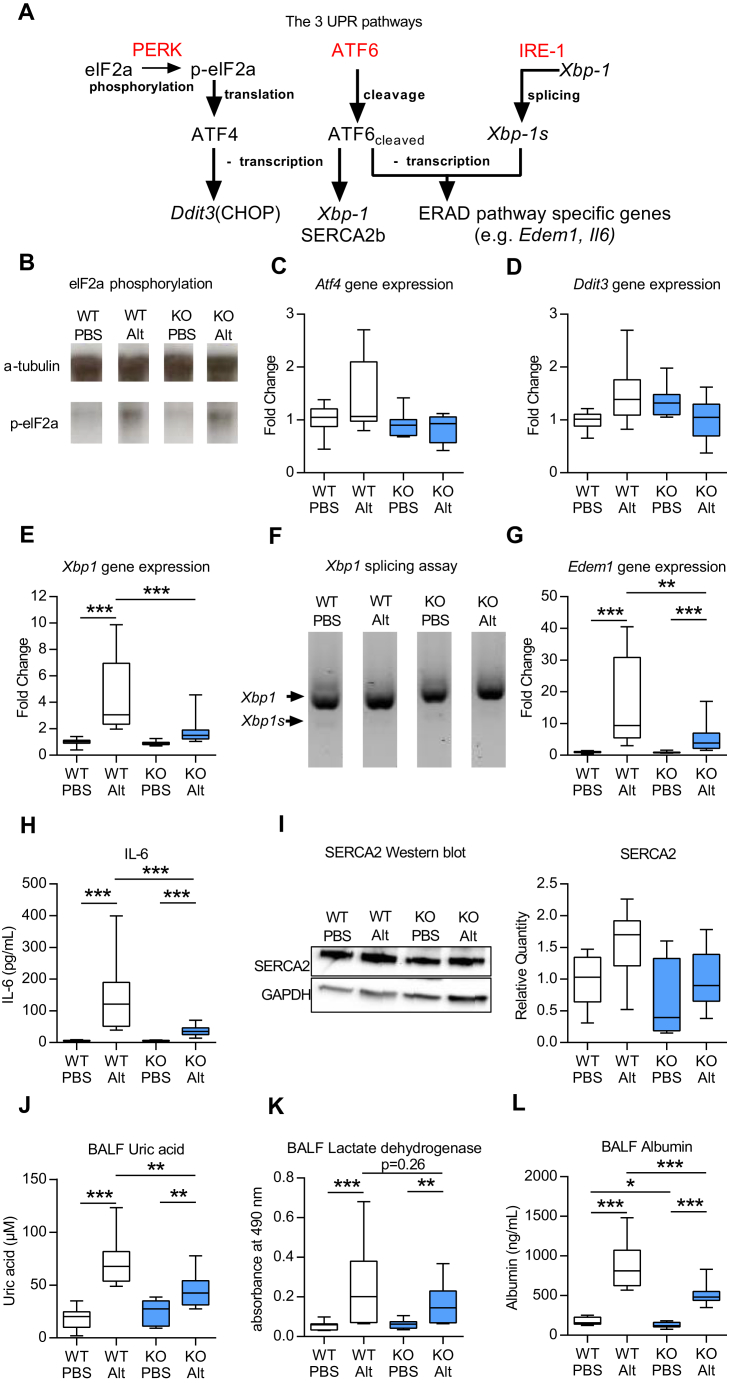
Ormdl3 KOMer mice are protected from developing Alternaria-induced AHR. A, Expression of Ormdl1 to Ormdl3 in lungs of naive WT and KO mice by quantitative PCR. B, Experimental plan (C-H). Lung function measured using the FlexiVent system. Dose-response curve to methacholine showing (Fig 1, C) airway resistance, (Fig 1, E) airway elastance, and (Fig 1, G) airway compliance. Airway resistance (Fig 1, D), airway elastance (Fig 1, F), and airway compliance (Fig 1, H) at 200 mg/mL methacholine. I, Concentration-response curve of tracheas toward increasing concentrations of methacholine. J, EC50 values of contraction responses. Alt, Alternaria; BL, baseline; i.n., intranasal; Data were collected from 2 individually performed experiments. N = 8-12 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005.
Following a single dose of the fungal allergen Alternaria, Ormdl3 mRNA expression has been shown to be induced in bronchial epithelial cells.5 To investigate the functional role of ORMDL3 during the development of asthma, we therefore developed a chronic Alternaria-induced AAD model (Fig 1, B). In WT mice, Alternaria induced the hallmark features of asthma, including AHR (Fig 1, C-H), lung and airway inflammation, as well as eosinophilia (Fig 2, A-D). IgE concentrations were also augmented following allergen challenge (Fig 2, E and F). We also observed an increase in pulmonary IL-13+ T cells (TH2) and type 2 innate lymphoid cells, enumerated by flow cytometry (Fig 3, A and B). Concomitant with the elevated AHR, pulmonary levels of IL-13 were increased (Fig 3, C).
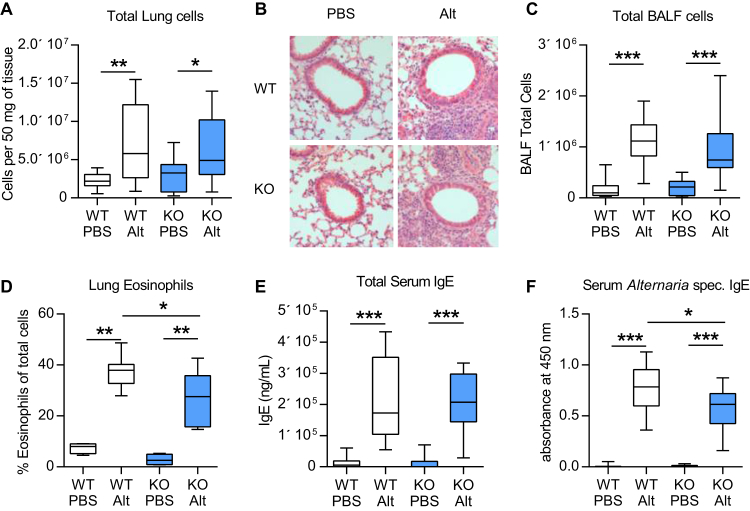
Fig 2.
Analysis of Alternaria-induced AAD in WT and Ormdl3 KOMer mice. A, Lung total cell counts. B,Alternaria-induced pulmonary inflammation in tissue sections (H&E staining). C, BALF total cell counts. D, Eosinophils expressed as the percentage SiglecF+ CD11c− CD68− lung cells. E, Total IgE concentration. F,Alternaria-specific IgE concentration. Representative photomicrographs are shown. Original magnification ×20. Alt, Alternaria; H&E, hematoxylin & eosin. Data were collected from 2 individually performed experiments. N = 8-16 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005.
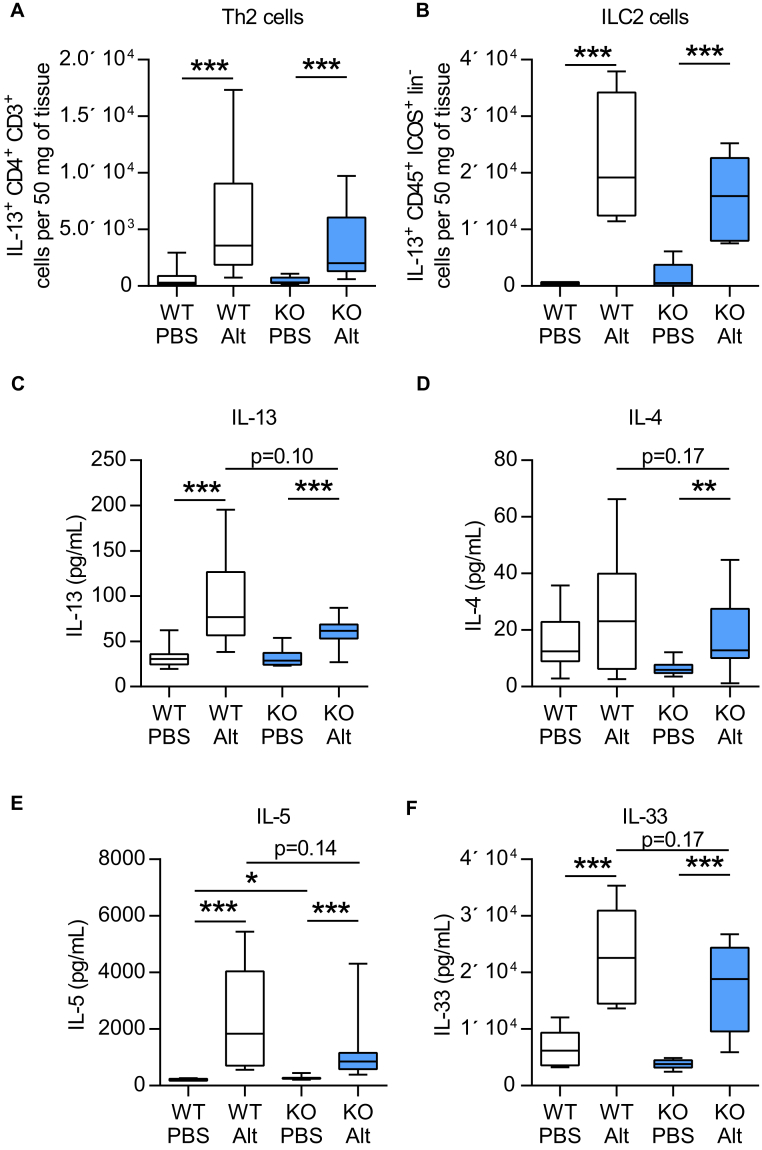
Fig 3.
ORMDL3 does not influence Alternaria-induced type 2 inflammation or mediators. A, Pulmonary TH2 cells (IL-13+ CD4+ CD3+). B, ILC2 cells (IL-13+ ICOS+ CD45+ lineage− cells). IL-13 (C), IL-4 (D), IL-5 (E), and IL-33 (F) cytokine levels in lung tissue determined by ELISA. Alt, Alternaria. Data were collected from 2 individually performed experiments. N = 8-16 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. ILC2, Type 2 innate lymphoid cell. *P < .05, **P < .005, and ***P < .0005.
Mice deficient in Ormdl3 were protected from the development of Alternaria-induced airway resistance and elastance (Fig 1, C-F) and allergen-induced decreases in airway compliance (Fig 1, G and H). Changes in airway resistance result from altered contractility of the smooth muscle surrounding the airways. Therefore, we chose to investigate whether intrinsic defects in smooth muscle cells could account for differences in lung function between WT and Ormdl3 KOMer mice. Tracheal rings from naive mice were isolated and the contraction responses toward increasing methacholine concentrations were compared (Fig 1, I). The methacholine EC50 value was significantly reduced in mice lacking ORMDL3 (Fig 1, J), indicating that ORMDL3 plays a role in airway smooth muscle contractility.
To further dissect the mechanisms that may account for the reduced Alternaria-induced AHR in Ormdl3 KOMer mice, we profiled the inflammatory cells and mediators present in the lung. Alternaria induced cell recruitment to the lung (Fig 2, A and B) and bronchoalveolar lavage (Fig 2, C) was not affected by pulmonary expression of ORMDL3, although the proportion of eosinophils was minimally but statistically significantly reduced (38% vs 28% SiglecF+CD11c−CD68− cells of total cells) in allergen-exposed Ormdl3 KO miceMer (Fig 2, D). Class switching to the allergy-associated IgE was unaffected by the loss of ORMDL3 (Fig 2, E), although there was a modest decrease in the levels of Alternaria-specific IgE (0.78OD vs 0.61OD) (Fig 2, F) in Ormdl3 KO miceMer.
The number of TH2 (CD4+IL-13+) cells and type 2 innate lymphoid cells (LinnegICOS+IL-13+) recruited to the lung (Fig 3, A and B) and bronchoalveolar lavage (data not shown) was not different between Ormdl3-sufficient and Ormdl3-deficient mice. Despite the reduction in AHR in mice lacking ORMDL3, these mice responded to Alternaria exposure with increased levels of pulmonary TH2 cytokines including IL-13, IL-4, and IL-5 in the lung (Fig 3, C-E). Concentrations of these cytokines were less than those observed in WT mice, but the reduction was not statistically significant. A small but significant increase in baseline levels of IL-5 was also noted (204 pg/mL vs 253 pg/mL) in the lung tissue (Fig 3, E). Similarly, Alternaria-induced increases in the levels of the pulmonary alarmin IL-33 were marginally reduced in Ormdl3 KOMer mice (Fig 3, F).
Thus, analysis of Ormdl3 KOMer mice reveal that lack of ORMDL3 protects against the development of fungal allergen–induced AHR, even though the mice are able to mount a robust TH2 response in the lung and periphery.
Ormdl3 KO mice have reduced cellular stress responses to Alternaria
In vitro and in vivo studies have suggested that ORMDL3 facilitates the UPR.5, 9, 14 Therefore, to further investigate how Ormdl3 deficiency, which does not affect type 2 immune responses in response to chronic Alternaria exposure, results in protection from allergen-induced AHR, the UPR was assessed. We specifically examined whether Alternaria promotes ER stress, and consequently UPR signaling in the lung, and more importantly, whether the magnitude of this response is altered in Ormdl3 KO mice. ER stress leads to detachment of the ER chaperone binding immunoglobulin protein from the 3 key ER stress proteins PERK, ATF6, and inositol requiring ER-to-nucleus signal kinase (IRE)-1α, thus initiating 3 parallel pathways of the UPR signaling cascade (Fig 4, A).11, 12 IRE-1α and PERK homodimerise and become activated following autophosphorylation of their respective cytoplasmic domains.11 ATF6 is cleaved by proteases in the Golgi complex and subsequently translocates to the nucleus and acts as a transcription factor.18 Activated PERK phosphorylates eIF2α, resulting in translational arrest of most proteins. Treatment of mice with Alternaria resulted in phosphorylation of eIF2α; however, generation of p-eIF2α was not modulated by ORMDL3 (Fig 4, B). Phosphorylation of eIF2α also selectively induces the transcription factor ATF4,19 which subsequently increases the expression of DNA-damage-inducible transcript 3, the gene encoding CCAAT/enhancer-binding protein-homologous protein.20 Baseline expression of Atf4 mRNA and DNA-damage-inducible transcript 3 was similar between WT and Ormdl3 KOMer mice and was not modulated by exposure of mice to Alternaria (Fig 4, C and D).
Fig 4.
Ormdl3 KOMer mice have diminished cellular stress responses to Alternaria. A, Schematic showing major components of the UPR. B, Western blot showing p-eIF2α. Specific quantitative PCR analyses were performed for Atf4(C), Ddit3(D), and total Xbp1(E). F, PCR of total and spliced Xbp1. G,Edem1 quantitative PCR. H, IL-6 determined by ELISA. I, Western blot showing SERCA2b. Uric acid (J), LDH (K), and albumin (L) levels in BALF. Alt, Alternaria; Ddit3, DNA-damage-inducible transcript 3; Edem1, ER degradation enhancer, mannosidase alpha-like 1. Data were collected from 2 individually performed experiments. N = 8-16 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005. For analysis of gene expression, Mann-Whitney statistical tests were performed between groups where a greater than 2-fold induction of median values was observed.
The ATF6- and IRE-1–mediated signaling cascades result in activation of the endoplasmic reticulum–associated protein degradation pathway. Acting as a transcription factor, cleaved ATF6 increases the expression of a second transcription factor X-box binding protein 1 (Xbp1).21 Interestingly, we observed an upregulation of total Xbp1 mRNA expression in WT mice in response to Alternaria (Fig 4, E). This was not observed in Ormdl3 KOMer mice. IRE-1α splices Xbp1 mRNA21; however, there was very little generation of the spliced Xbp1 gene product in any of the groups (Fig 4, F), indicating that Alternaria inhalation does not activate this pathway. Activation of ATF6 induces the transcription of genes that contribute to the clearance of unfolded/misfolded protein from the ER by binding to ER stress response elements. ER degradation enhancer, mannosidase alpha-like 1 (a protein degradation factor)22 is an example of an ATF6-inducible gene and its expression was increased in Alternaria-treated WT mice compared with PBS controls whereas transcription of this gene in response to allergen challenge in Ormdl3 KOMer mice was significantly reduced (Fig 4, G). The UPR is also linked to the production of several inflammatory cytokines and ATF6 binds to cAMP response elements promoting IL-6 mRNA transcription.23 Exposure of WT mice to Alternaria induced a robust IL-6 response in WT mice and this was significantly blunted in Ormdl3 KOMer mice (Fig 4, H). In vitro and in vivo studies have demonstrated that ORMDL3 overexpression results in increased expression of another ATF6 target gene, Atap2a2, that encodes SERCA2b.5, 9 However, using Ormdl3-deficient mice we show that ORMDL3 is not an absolute requirement for the expression of SERCA2b because basal and Alternaria-induced SERCA2b levels were not significantly different between WT and Ormdl3 KOMer mice (Fig 4, I). Thus, although the UPR is not affected by the absence of ORMDL3 under homeostatic conditions, ATF6 signaling as indicated by increased expression of ER degradation enhancer, mannosidase alpha-like 1 and IL-6 is significantly impaired in pathological conditions where ER stress is induced by exposure to Alternaria. Thus, the data indicate that ORMDL3 is involved in the regulation of key genes and proteins induced by ATF6 during ER stress.
There is also a UPR-independent branch of the ER stress response that regulates activation of the NLRP3 inflammasome.24 Alternaria exposure resulted in increased release of the damage-associated molecular pattern, uric acid. Concomitant with the reduced UPR in Ormdl3 KOMer mice, ORMDL3 deficiency also protected against UPR-independent cellular stress as evidenced by the significant reduction in uric acid released in these mice compared with WT animals (Fig 4, J). ORMDL3 specifically affects the ER stress pathway since levels of lactate dehydrogenase release from cells, a marker of nonspecific cellular damage, although a modest increase in response to Alternaria was not modulated by lack of ORMDL3 (Fig 4, K). Concurrent with the decrease in allergen-induced uric acid levels, the Ormdl3 KOMer mice also had significantly lower levels of bronchoalveolar lavage fluid (BALF) albumin following Alternaria challenge. This is indicative of maintained epithelial integrity and barrier function in Ormdl3 KOMer mice compared with WT animals (Fig 4, L). Further studies are needed to determine whether induction of cellular stress is an association or causally related to induction of AHR.
Overexpression of ORMDL3 promotes allergen-induced AHR
We next investigated the effect of overexpression of ORMDL3 in the lung using an AAV. Comparative experiments using different AAV serotypes (1, 2, 5, 6, 7, 8, 9, and rh10) encoding the marker gene EGFP were conducted. These revealed that intranasal treatment of neonatal mice in the first week of life with AAV9, in combination with neuraminidase to reveal galactose residues on the apical surface of conducting airway epithelial cells to increase AAV9 binding,25 resulted in optimal levels of EGFP expression in the lung (data not shown). Because of the lack of available ORMDL3-specific antibodies to track expression, an Ormdl3-EGFP gene fusion was introduced into the plasmid pZac2.1 to generate an AAV9 Ormdl3-EGFP virus (AAV Ormdl3). Five-day-old Balb/c mice were treated with vehicle (PBS), AAV EGFP, or AAV Ormdl3. At age 8 weeks, (adult) mice were exposed to Alternaria for 5 weeks and parameters of AAD were assessed (Fig 5, A). A lower dose of Alternaria was specifically chosen to determine any potential exacerbation of disease parameters in the mice overexpressing epithelial ORMDL3.
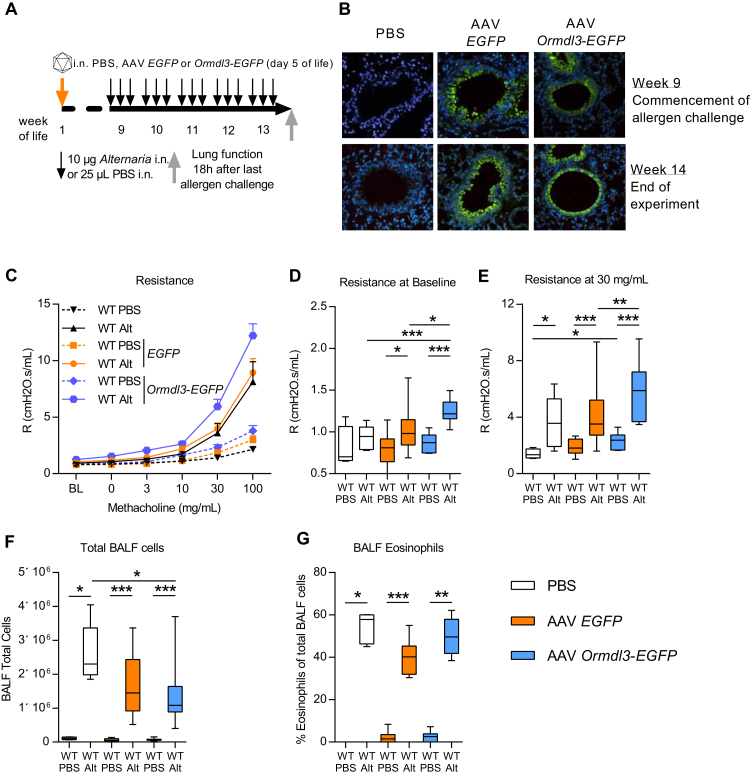
Fig 5.
ORMDL3 overexpression in airway epithelial cells enhances AHR. A, Experimental plan. B, GFP expression (staining green) in the lungs. C-E, Lung function measured using the FlexiVent system. Fig 5, C, Dose-response curve to methacholine showing airway resistance. Airway resistance at baseline (Fig 5, D) and 30 mg/mL methacholine (Fig 5, E). F, Total cells in the BALF. G, Eosinophils. Alt, Alternaria; Representative photomicrographs are shown. Original magnification ×20. Data were collected from 3 individually performed experiments. N = 8-12 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005.
Treatment of neonatal mice with AAV EGFP and AAV Ormdl3 resulted in robust levels of GFP expression primarily in bronchiolar epithelial cells and this expression persisted into adulthood (Fig 5, B). There was no effect of treating neonatal mice with the control AAV EGFP vector and these animals were phenotypically indistinguishable from PBS-treated littermate controls subsequently exposed to either PBS or Alternaria (Fig 5, C-G). At homeostasis, overexpression of ORMDL3 did not affect either parameters of lung function or the inflammatory profile of the mice (Fig 5, C-G). However, overexpression of ORMDL3 had a significant effect on Alternaria-induced AHR (Fig 5, C). At baseline (before the MCh challenge), Alternaria-exposed AAV Ormdl3 mice had significantly higher airway resistance than did allergen-exposed PBS or AAV EGFP-treated mice (Fig 5, D). Airway responsiveness to MCh challenge was also increased in the mice overexpressing ORMDL3 (Fig 5, E). In contrast, recruitment of cells to the airways was not affected by ORMDL3 overexpression (Fig 5, F and G). Alternaria-induced uric acid release was elevated in the ORMDL3-overexpressing mice although the increase was not statistically significant (data not shown).
Epithelial-specific ORMDL3 expression restores Alternaria-induced AAD
The previous series of experiments indicated that overexpression of ORMDL3 regulates lung function. Many of the recently identified asthma susceptibility genes including ORMDL3 are expressed in epithelial cells.3, 5 In WT mice, Ormdl3 is equally expressed by epithelial (EpCam+) and hemopoietic (CD45+) cells (Fig 6, A). To assess the particular contribution of epithelial-derived ORMDL3 expression to the generation of Alternaria-induced pathology, we reconstituted ORMDL3 expression in airway epithelial cells of Ormdl3 KOHar mice. Intranasal instillation of AAV Ormdl3 to neonatal mice resulted in expression almost exclusively in bronchial epithelial cells of the conducting airways as shown by immunofluorescence (Fig 6, B) and confirmed by quantitative PCR of sorted epithelial (EpCam+) and hemopoietic cells (CD45+) (Fig 6, C).
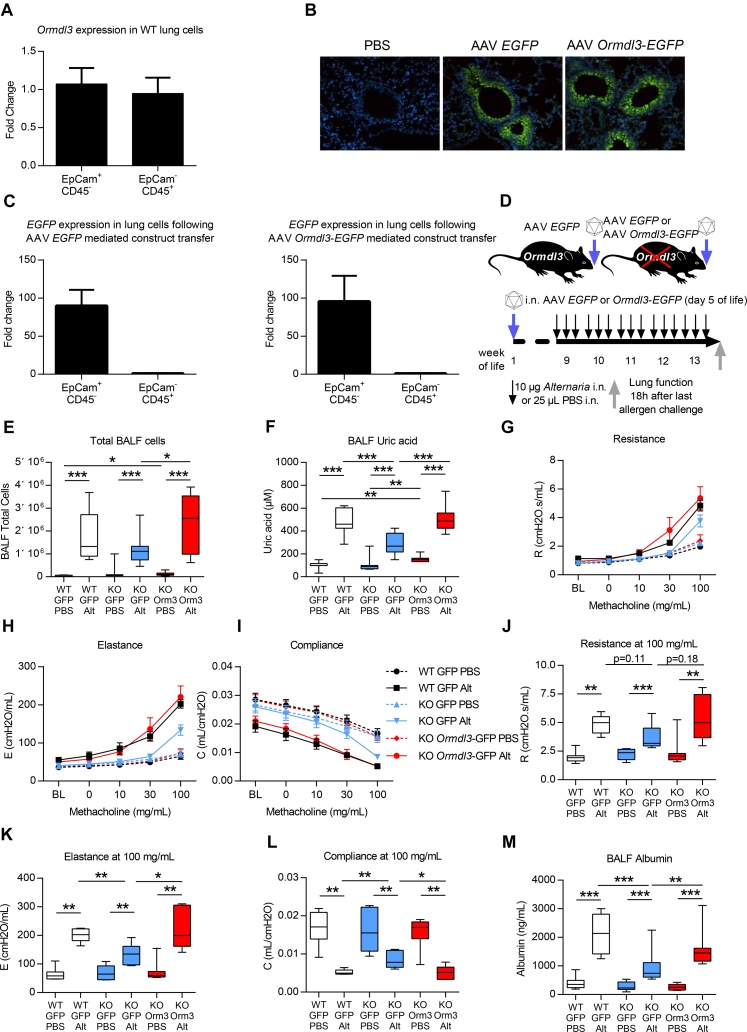
Fig 6.
Epithelial ORMDL3 expression drives susceptibility to Alternaria-induced AHR. A,Ormdl3 expression in epithelial (EpCam+) cells and leukocytes (CD45+) sorted from WT mice. B, GFP expression (staining green) in the lungs. C,EGFP expression in sorted epithelial cells and leukocytes from Ormdl3 KOHar mice following administration of either AAV EGFP or AAV Ormdl3. D, Experimental plan. E, Total cells in the BALF. F, Uric acid concentration. G-L, Lung function measured using the FlexiVent system. Airway resistance (Fig 6, G and J), airway elastance (Fig 6, H and K), and airway compliance (Fig 6, I and L). M, Albumin levels determined in BALF. Alt, Alternaria; i.n., intranasal. Representative photomicrographs are shown. Original magnification ×20. Data were collected from 2 individually performed experiments. N = 7-12 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005.
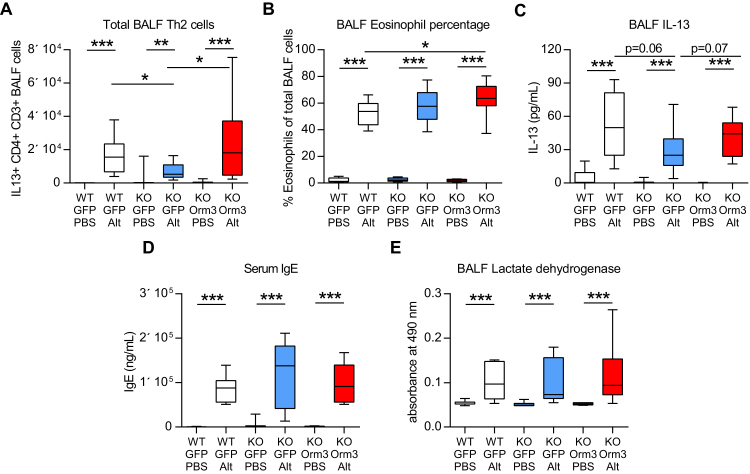
At age 8 weeks, WT, KOHar, and KOHar mice expressing ORMDL3 specifically in the airway epithelium were exposed to Alternaria for 5 weeks and parameters of AAD were assessed (Fig 6, D). Expression of epithelial ORMDL3 was associated with a small but statistically significant increase in total cells in the BALF (WT GFP PBS 6.5 × 104 vs KO Ormdl3 PBS 9.8 × 104) and uric acid (WT EGFP PBS 99 μM and KO EGFP PBS 90 μM vs KO Ormdl3 PBS 149 μM) in the airways but there was no other baseline phenotype in the absence of allergen challenge (Fig 6, D and E, and Fig E2). Restoration of epithelial ORMDL3 expression in the KOHar mice resulted in enhanced Alternaria- induced AHR that was equal in magnitude to that recorded in WT mice, implying that epithelial-derived ORMDL3 governs the deleterious Alternaria-induced changes in lung function (Fig 6, G-L). The total number of leukocytes recruited to the airway lumen following Alternaria challenge was increased in the KO mice expressing epithelial ORMDL3 compared with Ormdl3-deficient mice (Fig 6, E). Low-dose Alternaria revealed an effect on TH2 cells, with reduced recruitment in Ormdl3 KOHar mice (Fig E2, A). Epithelial expression of ORMDL3 in KOHar mice reestablished the allergen-induced increase in TH2 cell accumulation (Fig E2, A). There was also a small but significant increase in airway eosinophilia in the epithelial ORMDL3-expressing KOHar mice compared with WT mice (Fig E2, B). Congruent with the observed changes in type 2 inflammation, BALF IL-13 levels were reduced in Ormdl3 KOHar mice although the reduction was not statistically significant (Fig E2, C). IL-13 concentrations in Ormdl3 KOHar mice reconstituted with epithelial ORMDL3 were not different from those in WT mice. There was no effect of ORMDL3 on total IgE (Fig E2, D) or Alternaria-specific IgE levels in allergen-exposed mice (data not shown). Levels of the cell damage marker lactate dehydrogenase were similar in all groups of mice exposed to allergen (Fig E2, E), congruent with our previous observations. Viral expression of ORMDL3 in KOHar mice restored the uric acid response to Alternaria (Fig 6, F). In response to allergen, levels of BALF albumin were decreased in Ormdl3 KOHar mice compared with WT mice, indicative of increased epithelial integrity (Fig 6, M). However, in the KOHar mice reconstituted with epithelial ORMDL3, albumin levels were equivalent to those in WT mice (Fig 6, M). These data show that epithelial ORMDL3 governs the cellular stress response and AHR induced by inhaled Alternaria.
Fig E2.
Effect of epithelial ORMDL3 expression on type 2 inflammation. A, TH2 cells (IL-13+ CD4+ CD3+) in the BALF. B, Eosinophils expressed as the percentage SiglecF+ CD11c− CD68− in the BALF. C, IL-13 levels in the BALF. Serum IgE levels (D) and lactate dehydrogenase levels (E) in the BALF. Alt, Alternaria. Data were combined from 2 individually performed experiments. N = 7-12 mice per group. Box and whisker plots depict the median and interquartile range and minimum and maximum values. *P < .05, **P < .005, and ***P < .0005.
Discussion
GWAS have established a strong correlation between SNPs at the 17q21 locus and the development of asthma and ORMDL3 has been proposed as an asthma susceptibility gene. However, direct in vivo evidence for a pathophysiological function of ORMDL3 in a long-term model of AAD induced by a relevant aeroallergen associated with human disease is lacking.
ORMDL3 expression did not influence the number of TH2 cells or type 2 innate lymphoid cells recruited to the lung in response to Alternaria; however, eosinophilia was reduced in mice lacking ORMDL3. These data are in agreement with published literature demonstrating that ORMDL3 regulates eosinophil trafficking, recruitment, and degranulation.17 However, in our lower dose Alternaria exposure regime used in the epithelial ORMDL3 reconstitution experiments (Fig 6), we show that the total number of inflammatory cells recruited to the airway lumen are increased in mice expressing epithelial ORMDL3 compared with the global KO animals. The increased BAL cellularity was due to increased numbers of TH2 cells and eosinophils, indicating that epithelial ORMDL3 facilitates the trafficking of these cells to the airway lumen. However, the ORMDL3-dependent element of allergen-induced inflammation is overcome with higher doses of Alternaria. Levels of total IgE were unaffected by the loss of ORMDL3, indicating that ORMDL3 primarily affects innate rather than adaptive immunity in the generation of AAD.
Our data show that Ormdl3 KO mice are protected from the development of increased airway resistance, a function of smooth muscle contractility, following prolonged Alternaria exposure, implying that ORMDL3 is a critical driver of this aspect of AHR. Interestingly, although ORMDL3 is pivotal for the generation of Alternaria- induced AHR, the same phenotype was not observed in response to the aeroallergen house dust mite (data not shown). Ormdl3 KO mice develop AAD when exposed to house dust mite and the magnitude of AHR is equivalent to allergen-treated WT mice. This suggests that ORMDL3 mediates an Alternaria-specific activation of AAD. Sensitization to Alternaria has been implicated in severe asthma risk and fungal exposure is known to be associated with an increase in life-threatening exacerbations of the disease.26, 27 Intriguingly, SNPs in the chromosome 17q21 region in addition to being reproducibly associated with asthma have been linked to asthma exacerbations requiring hospitalization and/or oral steroids.28, 29
Mice globally overexpressing ORMDL3 have previously been shown to have elevated levels of SERCA2b, which transports calcium from the cytosol to the ER.5, 9 However, expression of SERCA2b is not solely dependent on the presence of ORMDL3 because levels were not altered in Ormdl3-deficient mice. ORMDL3 has also been shown to inhibit the activity of SERCA2b and this modulatory effect is suggested to result from direct association of the 2 proteins.14 The reduced sensitivity of the smooth muscle from Ormdl3 KO mice to pharmacologically induced contraction may therefore be due to decreased intracellular calcium concentrations as a result of increased SERCA2b-mediated uptake to the ER in the absence of ORMDL3, and reduced activity of SERCA in airway smooth muscle is thought to contribute to airway remodeling in those with moderate/severe asthma.30 ORMDL3, which has been shown to be induced by Alternaria, also regulates ceramide biosynthesis, and high-level expression of ORMDL3 increases ceramide production.8 Allergen induces elevated ceramide levels with concomitant AHR, which is ablated in mice treated with the sphingosine analogue FTY 720, which reduces both ORMDL3 and ceramide levels.8 Thus, the reduced smooth muscle contractility measured in Ormdl3 KO mice may also result from dysregulated lipid homeostasis.
We show that ORMDL3 expression specifically in bronchiolar epithelial cells restores the Alternaria-induced increase in AHR, suggesting that although ORMDL3 may have a direct effect on airway smooth muscle, it can also act indirectly. Uric acid levels are increased in patients with asthma following segmental allergen challenge and treatment of HDM-challenged mice with uricase ablates allergen-induced AHR.31 We have shown that Alternaria-induced secretion of uric acid into the airways is blunted in Ormdl3 KO mice. Conversely, viral-mediated expression of ORMDL3 in airway epithelial cells of KO mice reinstated the uric acid response to allergen challenge, clearly demonstrating the importance of epithelial-derived ORMDL3 in mediating Alternaria-induced release of damage-associated molecular patterns. Interestingly, uric acid is released early in the immune response to protease allergens,32 and levels can be used as a biomarker for the severity of asthma exacerbations in patients.33 Epithelial cells have an active uric acid transport system, and basal secretion of uric acid by human airway epithelial cells has been demonstrated.34 Uric acid is known to have direct effects on smooth muscle cells including stimulating endothelin-1 expression and increased intracellular calcium concentrations, which can induce smooth muscle contraction.35, 36 Thus, epithelial-expressed ORMDL3 can modulate lung function via its effects on uric acid release induced by allergen inhalation.
Many of the recently identified asthma susceptibility genes, including ORMDL3, are known to be expressed in epithelial cells,3, 5 and disrupted airway epithelial barrier function is thought to be a critical controller of disease induction.2 Using Ormdl3 KO mice, we can infer that this protein has a vital role in the epithelial response to allergen and dysregulated expression of ORMDL3 likely contributes to a reduction in epithelial integrity as demonstrated by the increase in bronchoalveolar lavage albumin levels in ORMDL3-sufficient mice compared with KO animals. In KO mice expressing epithelial ORMDL3, Alternaria-induced increases in luminal albumin were restored, suggesting that epithelial ORMDL3 governs allergen-induced loss of epithelial integrity.
Another branch of the ER stress response is the UPR. The protein sensors PERK, IRE-1α, and ATF6 respond to ER stress by increasing the expression of proteins such as binding immunoglobulin protein and CCAAT/enhancer-binding protein-homologous protein,37 which have been shown to be elevated in patients with asthma and in a murine model of neutrophilic, steroid-resistant AAD induced by LPS/ovalbumin. These markers are positively correlated with AHR and therapeutic treatment of mice with 4-phenylbutyric acid, a potent ER stress inhibitor, has been shown to reduce AHR, implying that activation of the UPR, which is dependent on ORMDL3, is intimately linked with lung function. In the lung, ORMDL3 activates the UPR via the ATF6 pathway, increasing transcription of endoplasmic reticulum–associated protein degradation pathway-specific genes (Edem-1) and regulating the expression of IL-6, which has been implicated in the pathogenesis of asthma.13
It is not known how the levels of ORMDL3 expression achieved by AAV-mediated gene transfer to the airway epithelium compare with the in vivo levels in WT mice due to the lack of specific reagents to differentiate between ORMDL1, 2, and 3 isoforms because of the high level of homology between these proteins, which we have shown directly using KO mice. Despite this, the data clearly demonstrate that epithelial ORMDL3 is essential for the generation of AAD and GWAS indicate that only the ORMDL3 isoform is associated with asthma susceptibility. ORMDL3 has a cAMP response element (CRE) in the promoter region and protein kinase A–dependent cAMP response element binding protein phosphorylation results in binding to the cAMP response element and initiation of ORMDL3 transcription.38 ORMDL3 activates ATF6, increasing the transcription of cAMP response element responsive genes; thus, ORMDL3 may mediate its own expression via a positive feedback loop. In addition, Alternariol, a mycotoxin of Alternaria, increases cAMP response element binding protein binding to CREs39 and this may be the mechanism by which Alternaria increases ORMDL3 expression.
Specific clinical asthma phenotypes (recurrent severe exacerbations) have previously been shown to be associated with distinct genotypes (cadherin regulated family member 3, locus).40 Stratifying patients with severe asthma into 2 groups on the basis of their response to fungal exposure reveals that the patients who respond to fungal allergens have earlier onset symptoms and require more oral steroid therapy than do those who do not and experience a greater number of asthma exacerbations.41, 42, 43 GWAS data predict that elevated ORMDL3 expression increases the risk of severe asthma and asthma exacerbations; thus, given the association between ORMDL3 and Alternaria-induced AHR revealed in the present study, it would be pertinent to determine whether SNPs in the 17q21 locus are associated with severe fungal-associated asthma phenotypes. With the current emphasis on personalized medicine, confirmation of the mechanisms and therapeutic targets in these specific patients could offer tailored treatments for this group of patients with asthma who frequently present with severe disease and are resistant to current steroid treatments. The ER stress inhibitor sodium phenylbutyrate (4-phenylbutyric acid) is licensed for oral administration and used therapeutically in the management of inherited urea cycle disorders.44 Because of its additional chaperone function it would be of interest to determine whether this drug also has efficacy in patients with asthma carrying genetic variations in the ORMDL3 locus.
In conclusion, ORMDL3 is a crucial regulator of smooth muscle contractility and subsequent AHR, acting potentially via cellular stress pathways, the UPR, and uric acid release. Modulation of cellular stress may represent a novel therapeutic avenue for the treatment of asthma.
Key messages.
-
•
In response to fungal exposure, Ormdl3 mediates the ATF6-dependent pathway of the UPR and uric acid release.
-
•
The cellular stress promoted by Ormdl3 orchestrates AHR during allergic immune responses.
-
•
Epithelial ORMDL3 is crucial for the induction of airway resistance.
Acknowledgments
We thank Lorraine Lawrence for conducting histological sectioning and Gaelle Herledan and Tom Shea for excellent technical assistance. Dr Mark Birrell and Prof Maria Belvisi kindly allowed us access to their organ bath equipment. In addition, we acknowledge the South Kensington Campus Flow Cytometry Facility at Imperial College for assistance with flow cytometry experiments. We thank Merck for the kind gift of Ormdl3 KO mice and the staff at Harwell and Taconic for animal husbandry support. Some graphics were created using layouts created by Motifolio Inc.
Footnotes
This study was funded by the Wellcome Trust (grant nos. 087618/Z/08/Z, 096964/Z/11/Z, and 097117/Z/11/Z), but it had no input to study design; collection, analysis, or interpretation of data; writing of the report, or the decision to submit the report for publication.
Disclosure of potential conflict of interest: S. Loeser has received a grant from the Wellcome Trust. S. A. Walker has received a grant from Medical Research Council/Asthma UK Centre for Asthma and Allergic Mechanisms. C. M. Lloyd has received grants from the Wellcome Trust and Janssen Inc. The rest of the authors declare that they have no relevant conflicts of interest.
Methods
RT-PCR
Lung tissue samples were homogenized in TRIzol solution (Life Technologies, Warrington, UK) followed by addition of chloroform to extract nucleic acids. RNA was isolated using the RNeasy Mini Kit (Qiagen, Manchester, UK), including an on-column DNase digest with an RNase-free DNase set (Qiagen) as per manufacturer's instructions. cDNA was amplified with random primers and the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer's instructions.
Real-time RT-PCR reactions were performed on a ViiA 7 Real-time PCR System (Life Technologies) in conjunction with TaqMan Fast Advanced Master Mix (Life Technologies) and following TaqMan Gene Expression Assays (Life Technologies): Atf4 (Mm00515325_g1), DNA-damage-inducible transcript 3 (Mm01135937_g1), ER degradation enhancer, mannosidase alpha-like 1 (Mm00551797_m1), Xbp1 (Mm00457357_m1), Gapdh (Mm99999915_g1), and Hprt (Mm01545399_m1). Expression of each target gene was normalized to the arithmetic mean of the 2 housekeeping genes Gapdh and Hprt. Data were then expressed as fold-change of relative mRNA expression in experimental groups versus naive WT animals.
Xbp-1 and spliced Xbp-1 (Xbp-1s) transcripts were amplified by PCR (forward primer: ACACGCTTGGGAATGGACAC; reverse primer: CCATGGGAAGATGTTCTGGG). The products were separated using a 4% agarose gel (Ultrapure agarose; Invitrogen, Waltham, Mass).
Isolation and sorting of CD45+ EpCam− and CD45− EpCam+ cells
Airway epithelial cells were sorted as previously reported,E1 with minor amendments. Lungs were perfused with ice-cold PBS through the right cardiac ventricle. Dissociation solution (1.5 mL) (Dulbecco modified Eagle medium/F12 [no phenol red; Life Technologies]) substituted with 25 mM HEPES (Sigma-Aldrich, Dorset, UK), penicillin/streptomycin, and 5 mg/mL dispase II (Sigma-Aldrich) was injected into the lungs through the trachea using a 1-mL syringe, followed by 0.5 mL of 1% low gelling temperature agarose (Sigma-Aldrich). The trachea was consequently closed using a hemostat and the chest was cooled down with ice to facilitate agarose solidification. The lung lobes were then harvested and incubated at 37°C for 1 hour in 5-mL dissociation solution under slight agitation. The digested tissue was passed through a 100-μm cell strainer (BD, Oxford, UK), cells were precipitated by centrifugation, washed through a 100-μm cell strainer for a second time to remove any remaining, solidified agarose, and incubated at room temperature for 10 minutes in Dulbecco modified Eagle medium/F12/25 mM HEPES in the presence of 50 μg/mL DNase I (Roche, Welwyn Garden City, UK). Erythrocytes were lysed and the cell suspension was extracellularly stained in sorting buffer (1% BSA in PBS) with phycoerythrin-conjugated EpCam (eBioscience, Cheshire, UK) and Brilliant Violet 421–conjugated CD45 (Biolegend) antimouse antibodies and sorted using a BD FACSAria III cell sorter. QIAshredder and RNeasy micro kits (Qiagen) were used in combination to isolate RNA. cDNA was prepared using the SuperScript VILO Master Mix (ThermoFisher, Waltham, Mass). RNA/cDNA preparation steps were performed according to manufacturers' instructions. Transcript levels of EGFP were assessed by quantitative RT-PCR using EGFP-targeting Taqman gene expression assays (Mr04329676_mr; Thermo Fisher). Gapdh served as the reference gene.
Immunofluorescence
Paraffin sections were stained sequentially with rabbit anti-GFP antibody (Abcam, Cambridge, UK), biotinylated goat anti-rabbit (Jackson, Suffolk, UK), and streptavidin AlexaFluor488 (Molecular probes, Invitrogen). Tissue sections were visualized and images overlayed using a LeicaDM2500 microscope and QWin software (Leica Microsystems [UK] Ltd, Milton Keynes, UK).
Western blot
Lung tissue was homogenized at 50 mg/mL tissue in RIPA buffer (Sigma, Dorset, UK) followed by centrifugation at 13,000 rpm for 10 minutes at 4°C. The resultant supernatant was used for Western blot. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% (w/v) nonfat milk in TRIS-buffered saline with 0.05% tween-20. The following primary antibodies were used: rabbit monoclonal anti-SERCA2 (Abcam), rabbit polyclonal anti-GAPDH (Millipore, Watford, UK), rabbit polyclonal anti-GAPDH Phospho-eIF2α (Ser51) (Cell Signaling Technology, Leiden, The Netherlands), and rabbit monoclonal anti-alpha tubulin antibody (Abcam).
Flow cytometry
BALF recovery, lung tissue digest, cell counting, and differential cell counting were performed as described previously.E2
Single-cell suspensions from lung tissue and bronchoalveolar lavage were stimulated with 40 ng/mL PMA (Sigma-Aldrich) and 3 μg/mL ionomycin (MERCK Millipore, Watford, UK) in RPMI/10% FCS containing 10 μg/mL Brefeldin A (Sigma-Aldrich) at 37°C, 0.5% CO2 for 3 hours. Cells were then washed and incubated for 20 minutes at room temperature with 10 μL normal rabbit serum (Sigma-Aldrich). After washing, cells were stained for extracellular antigens in 5% FCS/1% BSA in PBS for 30 minutes at 4°C. Cells were then washed, fixed, and permeabilized using Fix/Perm kit (eBioscience, Hatfield, UK) before being stained for intracellular antigens. All antibodies were purchased from eBioscience except anti-ICOS (Biolegend, London, UK). Acquisition was performed using Fortessa (BD) and analysis with FlowJo software (Flowjo, Ashland, Ala).
Mediator analysis
Lung tissue was homogenized in HBSS (Life Technologies) containing protease inhibitor cocktail (Roche). Lung tissue supernatant, bronchoalveolar lavage, and serum were analyzed by ELISA; IL-4 and IL-5 (BD PharMingen, Oxford, UK), IL-13 (Mouse Quantikine Kit [R&D Systems, Abingdon, UK]), IL-33 (Mouse IL-33 DuoSet ELISA, R&D Systems), and albumin (Bethyl Laboratories Inc, Montgomery, Tex). Paired antibodies for IgE (R&D Systems) were used to measure serum immunoglobulin levels. For determination of Alternaria-specific IgE levels, high-binding ELISA plates were coated with 50 μg/mL Alternaria extract. Uric acid levels were determined by Amplex Red uric acid assay (Life Technologies) and lactate dehydrogenase levels by a lactate dehydrogenase–based, in vitro toxicology assay kit (Sigma-Aldrich), according to the manufacturers' instructions.
In vitro measurement of airway smooth muscle function
Tracheal tissue was harvested from WT or Ormdl3 KOMer mice and bathed in Kreb's Henseleit solution warmed to 37°C and aerated with 95% O2/5% CO2. The tracheas were then cleared of extraneous tissue and split into 2 rings by cutting along the transverse plane. Tissues were then sutured to force-displacement transducers and placed in tissue baths. Following time to stabilize, tissues were challenged with acetylcholine (1 mM) 3 times to confirm viability and to standardize responses. Cumulative concentration-response curves (10 nM-1 mM) were then generated to methacholine or vehicle (Kreb's Henseleit solution). Agonist concentration-response curves were subsequently fitted by least-squares, nonlinear regression based on the Hill equation (Prism 5, GraphPad Software Inc). Mean EC50 values were calculated by averaging data from interpolation of response curves constructed for each individual tissue within a data set.
References
- 1.Wenzel S.E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 2.Holgate S.T. The sentinel role of the airway epithelium in asthma pathogenesis. Immunol Rev. 2011;242:205–219. doi: 10.1111/j.1600-065X.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 4.Hjelmqvist L., Tuson M., Marfany G., Herrero E., Balcells S., Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-research0027. RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller M., Tam A.B., Cho J.Y., Doherty T.A., Pham A., Khorram N. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. PNAS. 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worgall T.S., Veerappan A., Sung B., Kim B.I., Weiner E., Bholah R. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5:186ra67. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 8.Oyeniran C., Sturgill J.L., Hait N.C., Huang W.C., Avni D., Maceyka M. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J Allergy Clin Immunol. 2015;136:1035–1046.e6. doi: 10.1016/j.jaci.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller M., Rosenthal P., Beppu A., Mueller J.L., Hoffman H.M., Tam A.B. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192:3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodish H.F., Kong N., Wikstrom L. Calcium is required for folding of newly made subunits of the asialoglycoprotein receptor within the endoplasmic reticulum. J Biol Chem. 1992;267:12753–12760. [PubMed] [Google Scholar]
- 11.Bertolotti A., Zhang Y., Hendershot L.M., Harding H.P., Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 12.Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 13.Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 14.Cantero-Recasens G., Fandos C., Rubio-Moscardo F., Valverde M.A., Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 15.Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snelgrove R.J., Gregory L.G., Peiro T., Akthar S., Campbell G.A., Walker S.A. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol. 2014;134:583–592.e6. doi: 10.1016/j.jaci.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha S.G., Ge X.N., Bahaie N.S., Kang B.N., Rao A., Rao S.P. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd D.J., Lee A.-H., Glimcher L.H. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 19.Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 20.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 22.Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y., Porter K., Parameswaran N., Bae H.K., Pestka J.J. Role of GRP78/BiP degradation and ER stress in deoxynivalenol-induced interleukin-6 upregulation in the macrophage. Toxicol Sci. 2009;109:247–255. doi: 10.1093/toxsci/kfp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menu P., Mayor A., Zhou R., Tardivel A., Ichijo H., Mori K. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis. 2012;3:e261. doi: 10.1038/cddis.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell C.L., Vandenberghe L.H., Bell P., Limberis M.P., Gao G.P., Van Vliet K. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dales R.E., Cakmak S., Burnett R.T., Judek S., Coates F., Brook J.R. Influence of ambient fungal spores on emergency visits for asthma to a regional children's hospital. Am J Respir Crit Care Med. 2000;162:2087–2090. doi: 10.1164/ajrccm.162.6.2001020. [DOI] [PubMed] [Google Scholar]
- 27.O'Driscoll B.R., Hopkinson L.C., Denning D.W. Mold sensitization is common amongst patients with severe asthma requiring multiple hospital admissions. BMC Pulm Med. 2005;5:4. doi: 10.1186/1471-2466-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisgaard H., Bonnelykke K., Sleiman P.M., Brasholt M., Chawes B., Kreiner-Moller E. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 29.Tavendale R., Macgregor D.F., Mukhopadhyay S., Palmer C.N. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–863. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Mahn K., Hirst S.J., Ying S., Holt M.R., Lavender P., Ojo O.O. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci. 2009;106:10775–10780. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kool M., Willart M.A., van Nimwegen M., Bergen I., Pouliot P., Virchow J.C. An unexpected role for uric acid as an inducer of T helper 2 cell immunity to inhaled antigens and inflammatory mediator of allergic asthma. Immunity. 2011;34:527–540. doi: 10.1016/j.immuni.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Hara K., Iijima K., Elias M.K., Seno S., Tojima I., Kobayashi T. Airway uric acid is a sensor of inhaled protease allergens and initiates type 2 immune responses in respiratory mucosa. J Immunol. 2014;192:4032–4042. doi: 10.4049/jimmunol.1400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L., Wan C., Wen F. An unexpected role for serum uric acid as a biomarker for severity of asthma exacerbation. Asian Pac J Allergy Immunol. 2014;32:93–99. doi: 10.12932/AP0337.32.1.2014. [DOI] [PubMed] [Google Scholar]
- 34.Gold M.J., Hiebert P.R., Park H.Y., Stefanowicz D., Le A., Starkey M.R. Mucosal production of uric acid by airway epithelial cells contributes to particulate matter-induced allergic sensitization. Mucosal Immunol. 2015;9:809–820. doi: 10.1038/mi.2015.104. [DOI] [PubMed] [Google Scholar]
- 35.Chao H.H., Liu J.C., Lin J.W., Chen C.H., Wu C.H., Cheng T.H. Uric acid stimulates endothelin-1 gene expression associated with NADPH oxidase in human aortic smooth muscle cells. Acta Pharmacol Sin. 2008;29:1301–1312. doi: 10.1111/j.1745-7254.2008.00877.x. [DOI] [PubMed] [Google Scholar]
- 36.Albertoni G., Schor N. Resveratrol inhibits the intracellular calcium increase and angiotensin/endothelin system activation induced by soluble uric acid in mesangial cells. Braz J Med Biol Res. 2015;48:51–56. doi: 10.1590/1414-431X20144032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S.R., Kim D.I., Kang M.R., Lee K.S., Park S.Y., Jeong J.S. Endoplasmic reticulum stress influences bronchial asthma pathogenesis by modulating nuclear factor kappaB activation. J Allergy Clin Immunol. 2013;132:1397–1408. doi: 10.1016/j.jaci.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang L.L., Jin R., Zhu L.H., Xu H.G., Li Y., Gao S. Promoter characterization and role of cAMP/PKA/CREB in the basal transcription of the mouse ORMDL3 gene. PLoS One. 2013;8:e60630. doi: 10.1371/journal.pone.0060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Liu K., Lu J., Ma J., Zhang X., Jiang Y. Alternariol induces DNA polymerase beta expression through the PKA-CREB signaling pathway. Int J Oncol. 2012;40:1923–1928. doi: 10.3892/ijo.2012.1398. [DOI] [PubMed] [Google Scholar]
- 40.Bonnelykke K., Sleiman P., Nielsen K., Kreiner-Moller E., Mercader J.M., Belgrave D. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 41.Downs S.H., Mitakakis T.Z., Marks G.B., Car N.G., Belousova E.G., Leuppi J.D. Clinical importance of Alternaria exposure in children. Am J Respir Crit Care Med. 2001;164:455–459. doi: 10.1164/ajrccm.164.3.2008042. [DOI] [PubMed] [Google Scholar]
- 42.Tham R., Dharmage S.C., Taylor P.E., Katelaris C.H., Vicendese D., Abramson M.J. Outdoor fungi and child asthma health service attendances. Pediatr Allergy Immunol. 2014;25:439–449. doi: 10.1111/pai.12257. [DOI] [PubMed] [Google Scholar]
- 43.Castanhinha S., Sherburn R., Walker S., Gupta A., Bossley C.J., Buckley J. Pediatric severe asthma with fungal sensitization is mediated by steroid-resistant IL-33. J Allergy Clin Immunol. 2015;136:312–322.e7. doi: 10.1016/j.jaci.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker V. Ammonia toxicity and its prevention in inherited defects of the urea cycle. Diabetes Obes Metab. 2009;11:823–835. doi: 10.1111/j.1463-1326.2009.01054.x. [DOI] [PubMed] [Google Scholar]
References
- Juncadella I.J., Kadl A., Sharma A.K., Shim Y.M., Hochreiter-Hufford A., Borish L. Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature. 2013;493:547–551. doi: 10.1038/nature11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelgrove R.J., Gregory L.G., Peiro T., Akthar S., Campbell G.A., Walker S.A. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol. 2014;134:583–592.e6. doi: 10.1016/j.jaci.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]