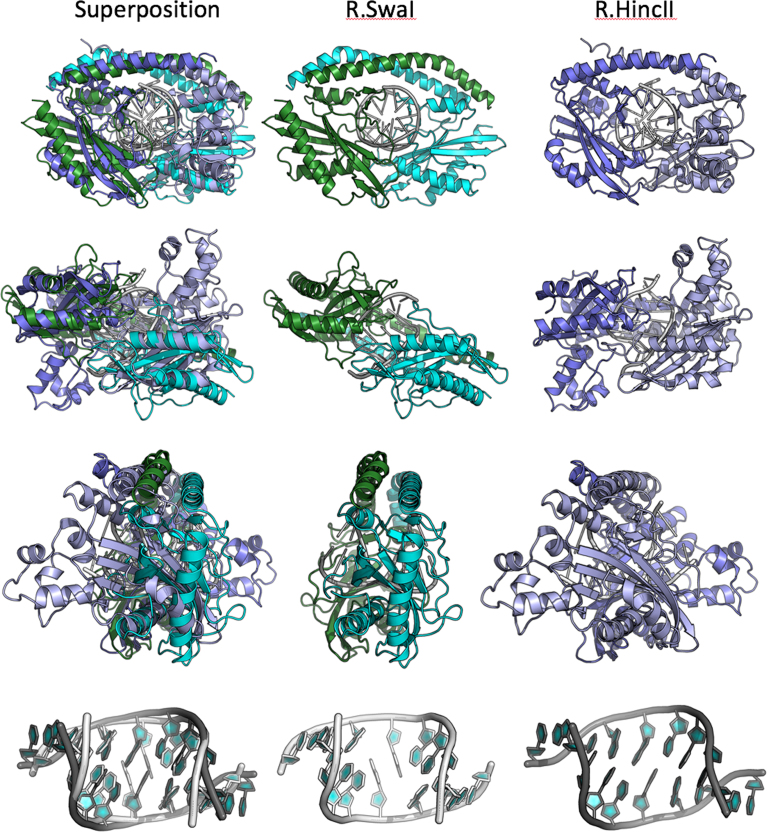
Figure 6.
Superposition and comparison of the DNA-bound structures of SwaI and HincII. The left panels in each row show the superposition of both structures; the middle and right panels show the same elements in their individual molecular complexes. Each superposition is calculated based on the core nuclease residues of the helix and 3 β-strands that comprise the core elements of the PD-(D/E)xK nuclease motif (see Figure 7). Note that while the nuclease domains superimpose relatively closely, that the corresponding orientations of additional protein structural elements (such as the domain-swapped helices) and the bound DNA differ by at least 10° from one another. The bottom row shows the superposition of the two bound DNA targets for SwaI and HincII. While the overall bend angle and positions of the flanking bases (at positions 2–4 in each half-site) are similar, the effect of DNA binding and bending on the central two base pairs are radically different. In SwaI the central A:T base pairs are completely disrupted, whereas in HincII (and in EcoRV, not shown) they remain in a bent (but still base-paired) conformation.