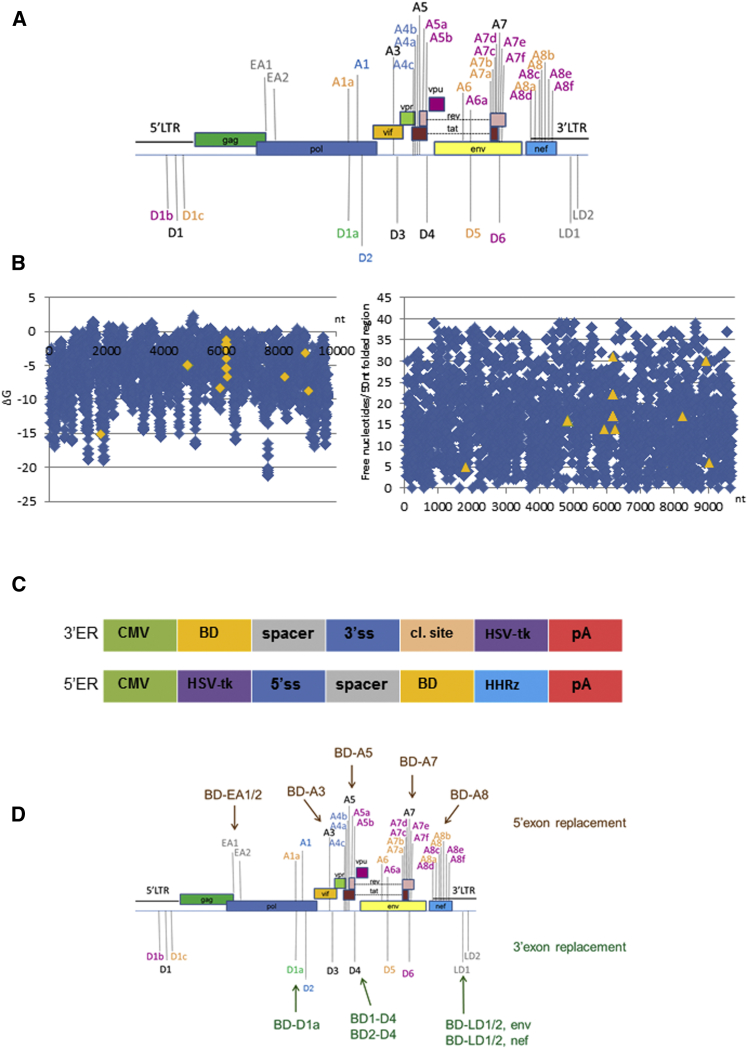
Figure 1.
In Silico Analysis of HIV Splicing and Splice-Site Target Determination for RNA Trans-Splicing
(A) Summary of HIV splice sites considered for targeting by RNA trans-splicing. Conserved splice donor/acceptor sites predicted by computer splice site predictions of pNL4.3 are shown in black.87 In green, previously reported cryptic splice donor site predicted by computer algorithms.41 In gray, late splice donor/early splice acceptor sites predicted in pNL4.3 by computer algorithms. In blue, conserved splice donor/acceptor sites not predicted in pNL4.3 by computer splice site predictions but previously published.87 In orange, previously reported cryptic splice sites not predicted in pNL4.3 by computer algorithms (A1a,41 A6,50, 51, 52 D5,50, 51 A7a, A7b,88 A8a, A8, A8b89). In purple, recently described splice donor and splice acceptor sites not predicted by computer algorithms,6 A5a.6, 89, 90 (B) Selection of binding domain regions using the software Foldanalyze (HUSAR). MFE analysis of pNL4.3 throughout the complete reverse complement pNL4.3 genome per 50 nt folded region (left). Number of free nucleotides per 50 nt folded region of the complete reverse complement pNL4.3 genome (right). Target regions of binding domains selected for further design are shown in orange. (C) Maps of CMV promoter-driven RNA trans-splicing cassettes. 3′ exon replacement (top) and 5′ exon replacement (bottom) cassettes are shown. BD, binding domain; spacer, spacer sequence; 3′ss, 3′ splice site domain; cl. site, P2A protein cleavage site91; HSV-tk, herpes simplex virus thymidine kinase gene; pA, polyA tail; HHRz, hammerhead ribozyme sequence. (D) Summary of selected HIV splice sites for targeting by 5′ exon replacement (top) or 3′ exon replacement (bottom). HIV splice site D4 was targeted with two different binding domain sequences (BD1-D4 and BD2-D4), and LD 1 and 2 (LD1 and LD2 respectively) were targeted with two BD-LD1/2 cassettes driven from either HIV Env or HIV Nef translational start sites.