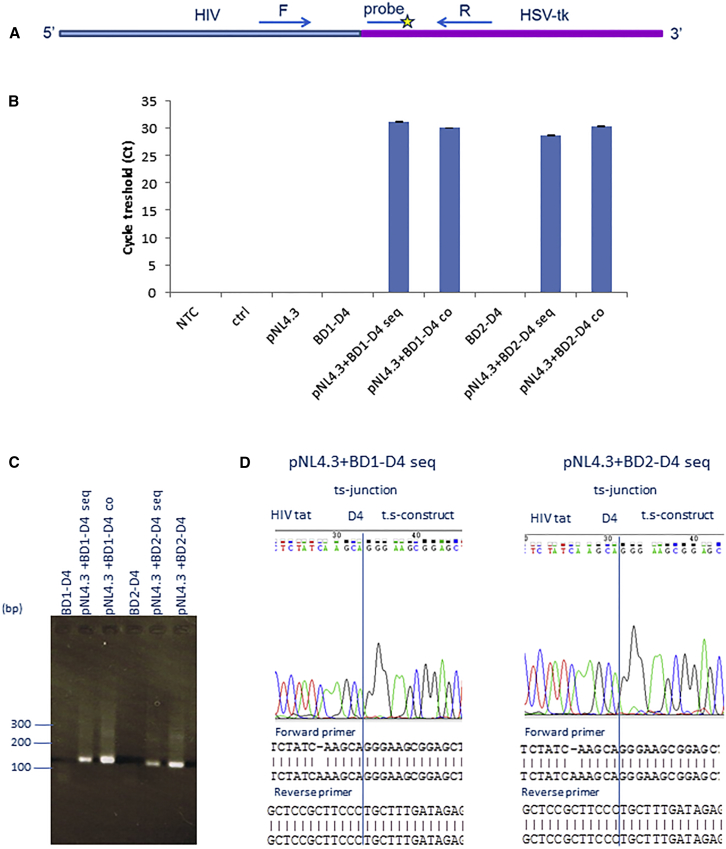
Figure 4.
Confirmation of RNA Trans-Splicing between HIV and 3′ Exon Replacement Constructs
(A) qRT-PCR primer and probe design to detect RNA trans-splicing amplicons. The forward primer is located in the HIV region and the reverse primer in the HSV-tk region. The PCR amplicon was detected with a probe in the HSV-tk region. (B) qRT-PCR data in cells transfected with the HIV proviral clone pNL4.3 followed by transfection with 3′ exon replacement construct BD1-D4 or BD2-D4. Cells were analyzed 48 hr post-transfection with the RNA trans-splicing constructs in sequentially transfected cells (seq) or co-transfected cells (co). (C) Confirmation of RNA trans-splicing by conventional PCR in sequentially (seq) or co-transfected cells (co). (D) Confirmation of RNA trans-splicing products by Sanger sequencing between HIV tat and HSV-tk constructs. BD1-D4 (left) and BD2-D4 (right). The error bars represent the standard deviation (STDEV) of average Ct values.