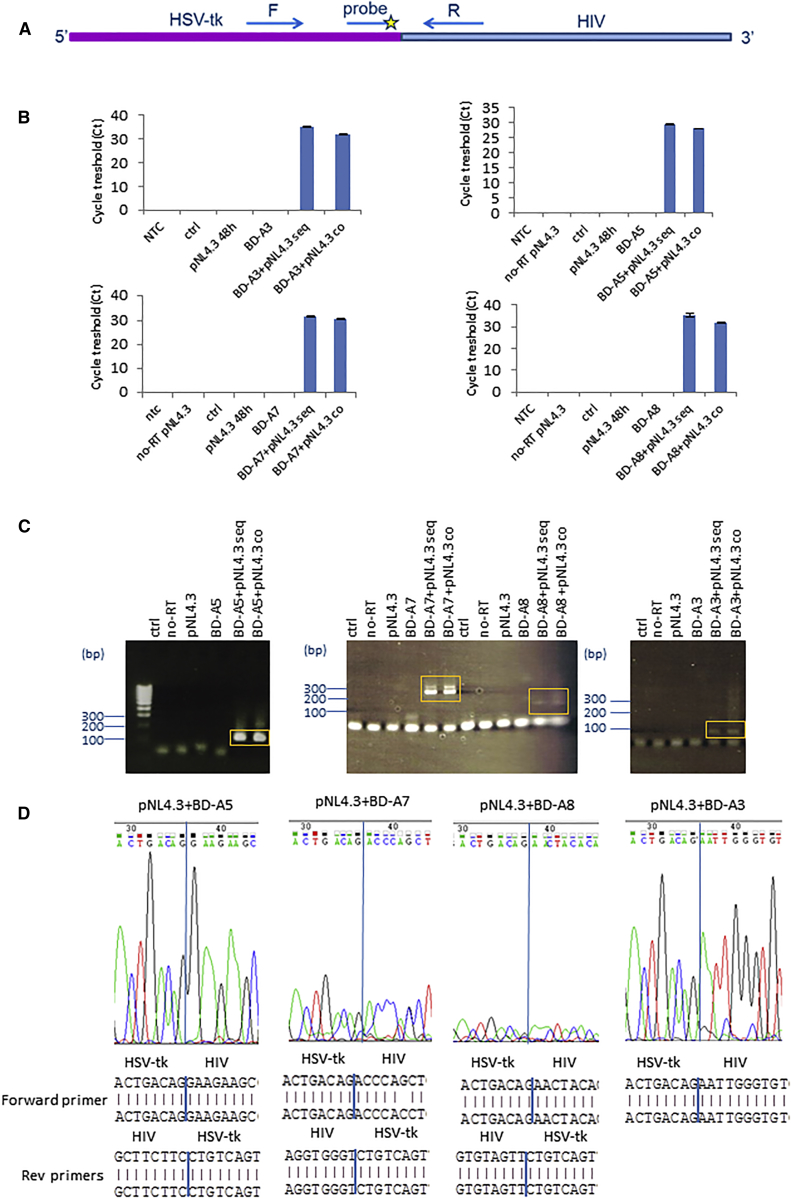
Figure 5.
Confirmation of RNA Trans-Splicing between HSV-tk and 5′ Exon Replacement Constructs
(A) qRT-PCR primer and probe design to detect RNA trans-splicing amplicons. The forward primer is located in the HSV-tk region and the reverse primer in the HIV region. The PCR amplicon was detected with a probe in the HIV region. (B) qRT-PCR data from cells transfected with the HIV proviral clone pNL4.3 followed by transfection with 5′ exon replacement constructs sequentially (seq) or after co-transfection of the two constructs (co). Top: data for BD-A3 and BD-A5 (left to right). Bottom: data for BD-A7 and BD-A8 (left to right). (C) Detection of HIV RNA trans-splicing products by conventional PCR. RNA trans-splicing was analyzed by with primers specific for BD-A5 (left), BD-A7 and BD-A8 (middle), and BD-A3 (right). PCR products highlighted in orange were PCR-purified and further analyzed by Sanger sequencing, as in (D). (D) Confirmation of RNA trans-splicing junctions by Sanger sequencing in sequentially transfected cells. Chromatographs showing sequence reads are included (top). PCR products as shown in (C) were analyzed by sequencing with a 5′ER forward primer and HIV-specific reverse primers. Sequencing data are shown from left to right for BD-A5, BD-A7, BD-A8, and BD-A3. RNA trans-splicing junctions are shown by blue lines on sequence chromatographs and sequence reads.