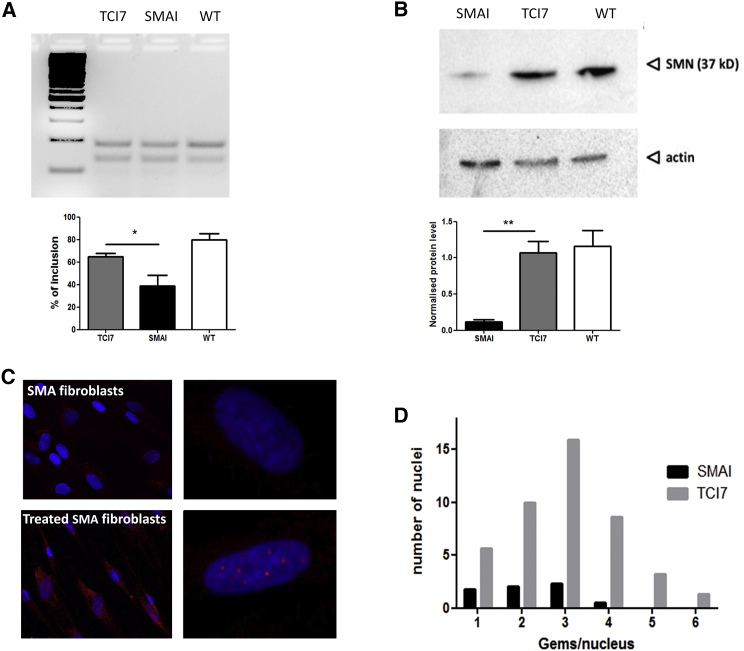
Figure 1.
Evaluation of In Vitro Efficiency of tcDNA TCI7
(A) Detection of the inclusion of exon 7 of SMN2 mRNA by RT-PCR (primers on exons 6 and 8) in patient SMA I fibroblasts. PCR products were visualized on 3% agarose gel (top) and quantified by ImageJ (bottom). TCI7, fibroblasts treated with tcDNA AONs targeting the ISS-N1; SMAI, untreated SMAI fibroblasts (GM03813 Coriell); and WT, healthy fibroblasts of the mother (GM03814 Coriell). (B) Detection of the SMN protein by western blot in fibroblasts and normalization with actin. Lane 1, untreated SMAI fibroblasts (GM03813 Coriell); lane 2, SMA fibroblasts treated with TCI7; lane 3, healthy fibroblasts of the mother (GM03814 Coriell). Error bars represent SD. *p < 0.05 and **p < 0.01. (C) Detection of the SMN protein by immunostaining in untreated SMA fibroblasts (top) and SMA fibroblasts transfected with TCI7 (bottom). Cells were immunostained with the polyclonal antibody H-SMN 195 (Santa Cruz). The red staining shows the expression of SMN protein in gems. Nuclei are labeled in blue (DAPI). Left: original magnification ×20; right: original magnification ×63. For magnification ×63, the apotome is used to see the gems in the nucleus. (D) Counting the number of gems stained positively for SMN in each nucleus (n = 100 nuclei). The black bars correspond to untreated SMA type I, and gray bars correspond to cells treated with TCI7.