Abstract
Accurate definition of genetic mutations causing Duchenne muscular dystrophy (DMD) has always been relevant in order to provide genetic counseling to patients and families, and helps to establish the prognosis in the case where the distinction between Duchenne, Becker, or intermediate muscular dystrophy is not obvious. As molecular treatments aimed at dystrophin restoration in DMD are increasingly available as commercialized drugs or within clinical trials, genetic diagnosis has become an indispensable tool in order to determine eligibility for these treatments. DMD patients in which multiplex ligation-dependent probe amplification (MLPA) or similar techniques show a deletion suitable to exon skipping of exons 44, 45, 51, or 53, may be currently treated with AONs targeting these exons, in the context of clinical trials, or, as is the case for exon 51 skipping in the United States, with the first commercialized drug (eteplirsen). Patients who test negative at MLPA, but in whom DMD gene sequencing shows a nonsense mutation, may be amenable for treatment with stop codon readthrough compounds such as ataluren. Novel molecular approaches such as CRISPR-Cas9 targeting of specific DMD mutations are still in the preclinical stages, but appear promising. In conclusion, an accurate genetic diagnosis represents the entrance into a new scenario of personalized medicine in DMD.
Key words: Duchenne muscular dystrophy, genetic diagnosis, exon skipping, stop codon readthrough, CRISPR-Cas9
Introduction
In 1987, the discovery that Duchenne muscular dystrophy (DMD) is caused by the absence of the protein dystrophin in skeletal muscle fibers (1) first provided a rationale for molecular treatments, aimed at restoring dystrophin expression. Shortly thereafter, the discovery that Becker muscular dystrophy (BMD) is a milder allelic form of dystrophinopathy, in which dystrophin is present but qualitatively altered and quantitatively reduced (2) suggested that clinical benefit may be provided to DMD patients, even with partial restoration of dystrophin expression. Despite the hope stirred by these seminal discoveries, after three decades such molecular treatments are not yet widely available to DMD patients. In fact, formidable obstacles still stand in the way of the translation of dystrophin-restoring treatments from proof-of-concept experiments to clinical trials and everyday clinical practice: for instance, because of several genetic and environmental factors, there is considerable variability in disease progression , which has made the design and interpretation of clinical trials more challenging (3, 4). Nevertheless, several molecular therapies for DMD have reached advanced stages of clinical experimentation, and the first drugs have recently reached the market through accelerated and provisional regulatory approvals (5, 6).
In the complex and swiftly changing scenario of experimental treatments for DMD, the expression "molecular therapy" requires a more precise definition. Dystrophin restoration may be achieved in two different ways: through genetic therapies, i.e. delivery of new genetic material, that is not naturally present in the patient's cells, such as a functioning copy of the DMD gene or transcript; or through molecular therapies, i.e. small or larger molecules (i.e. oligonucleotides, enzymes), that interact with the patients' own genetic material and transcription/translation machinery to restore the expression of viable dystrophin. Here, we intend to focus on molecular therapies, mainly for two reasons: first, molecular therapies have reached more advanced stages in human experimentation than genetic therapies for DMD, and are therefore of more immediate interest to neuromuscular clinicians; and second, they highlight the importance of reaching a well-defined molecular diagnosis, at the genomic level, in DMD. In fact, while genetic therapies would theoretically benefit any DMD patient, molecular therapies are an example of personalized therapies, as they are expected to work only in those patients who harbor specific mutations, targeted by the molecule of choice.
We will first briefly review the current standards in molecular diagnosis of DMD, and then the main molecular treatment strategies accessible to patients with different identified mutations: exon skipping through antisense oligonucleotides (AONs), and readthrough of premature termination codons through the small molecule ataluren. Recently, the prospect of editing patients' genomes using the CRISPR-Cas9 technology has opened a new exciting perspective in this field.
Genetic diagnosis of DMD
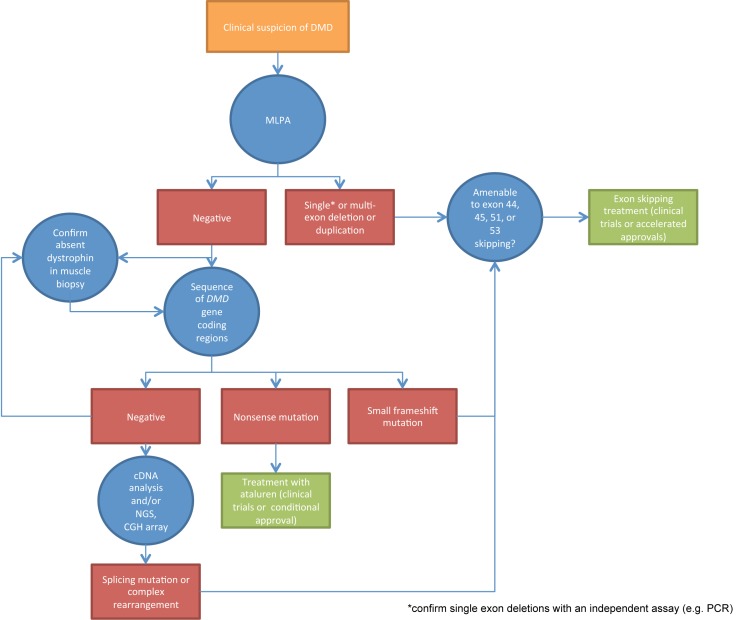
Typically, the diagnosis of DMD may be established based on the clinical picture and the finding of absent dystrophin expression in muscle by immunohistochemistry and/or immunoblot (7). However, internationally adopted guidelines (8, 9) indicate that an accurate characterization of the causative mutation at the genomic level is of paramount importance in order to provide genetic counseling to the family, establish genotype-phenotype correlations and prognosis, and, as we will show in the next paragraphs, assess eligibility for novel molecular treatments. The first molecular assay to be requested when DMD is suspected should be quantitative, i.e. provide a measure of the copy number of genomic regions corresponding to each of the 79 DMD exons. In fact, about 60 to 70% of DMD-causing mutations are large rearrangements (deletions or duplications) including one or more exons (10). These are readily identified in both affected males and female carriers by multiplex ligation-dependent probe amplification (MLPA) (11). In the case of single-exon deletions, results should be confirmed by an independent technique, as some small mutations may prevent probe hybridization and be misinterpreted as deletions of the corresponding exons. Alternative quantitative techniques include multiplex PCR and competitive genomic hybridization (CGH) arrays. Whenever quantitative assays cannot completely characterize the mutation, PCR amplification and sequencing of the coding DMD regions and flanking nucleotides allows the identification of single nucleotide substitutions, small sub-exonic rearrangements, and canonic splice-site mutations. While classic Sanger sequencing is still widely used, next generation sequencing (NGS) approaches are becoming increasingly accessible (9). In rare cases in which no mutation can be identified by sequencing of genomic DNA coding regions, DMD mRNA may be isolated from skeletal muscle tissue (either archived from a previous diagnostic biopsy, or obtained specifically for this purpose), reversetranscribed to cDNA, and sequenced. The localization of sequence anomalies in the transcript may point to deep intronic causative mutations that may be identified by targeted Sanger sequencing of intronic gDNA regions, or by NGS. The systematic application of this workflow in DMD patients (illustrated in Fig. 1) leads to the identification of a clear pathogenetic mutation in the vast majority of cases.
Figure 1.
Flow diagram of genetic diagnosis in DMD, highlighting indicated molecular assays (blue circles) and corresponding possible findings (red rectangles), leading to further assays, or, in some cases, to established amenability for molecular treatments (green rectangles).
Exon skipping with antisense oligonucleotides
The rationale for "exon skipping" is based on several facts: in about two thirds of cases, DMD is caused by large deletions involving one or more exons, and disrupting the DMD open reading frame (ORF); on the contrary, deletions that respect the ORF cause the milder allelic disease BMD (12); because of the DMD genomic structure, it is often possible to restore the ORF of an out-offrame deletion by splicing out just one exon, adjacent to the deletion borders at the 5' or 3' side, from the mature mRNA. Therefore, by targeting and inactivating specific exon splicing signals in the pre-mRNA with sequencespecific AONs, it is possible to obtain an internally deleted, yet in-frame DMD transcript, which is similar to transcripts naturally observed in BMD patients. These transcripts may then be translated into a viable, albeit internally deleted and quantitatively reduced, dystrophin protein, hopefully shifting the patient's clinical picture towards the milder BMD phenotype. The application of AON treatments to DMD and other genetic conditions became technically possible in the early 2000s, with the invention of chemically modified nucleotide backbones, which resisted to nucleases and had a favourable pharmacokinetic and pharmacodynamic profile in humans. These were 2'-O-methyl RNA phosphorothioate AONs (administered subcutaneously) and morpholino AONs (administered intravenously) (13). For both chemistries, first-in-human studies with local intramuscular injections of AONs showed promising dystrophin restoration in biopsies of injected muscles (14 ,15). The leading compounds targeted exon 51, whose skipping is predicted to restore the ORF in the highest portion of DMD patients (around 10-15%). Subsequent phase 2 dose-escalation studies (16, 17) were also promising, as they seemed to provide not only assurances of good tolerability of these compounds, but also biochemical evidence of dystrophin restoration and encouraging stabilization or improvement of some functional outcome measures. Unfortunately, in the years immediately following these exciting breakthroughs, the enthusiasm of the DMD community was thwarted. An international, multi-center phase 3 trial of drisapersen (clinicaltrials.gov NCT01254019), the 2'-Omethyl AON for the skipping of exon 51 developed by Biomarin/Prosensa, failed to achieve a significant clinical benefit (the results have not yet been published). While more partially encouraging data came from a phase 2b extension study (18), the company decided to discontinue the drisapersen development program. These events have triggered a lively debate (19, 20) about several controversial aspects of exon skipping treatments, such as the difficulty to accurately measure efficacy of treatments both at the molecular (21) and clinical level (22), and the amount of dystrophin needed to actually obtain a clinical benefit (23). As for eteplirsen, the morpholino AON for the skipping of exon 51 developed by Sarepta, phase 3 studies are still underway (NCT02255552), but interesting data have derived from a small but prolonged phase 2b extension study (24). Clinical data from this open-label study, also corroborated by comparison to a mutationmatched external natural history control group (25), seem to support stabilization of ambulatory function in 12 patients. Unfortunately, data regarding dystrophin quantification in several longitudinal muscle biopsies in the same study indicate less abundant protein expression increase than suggested by earlier studies, and have been the object of controversy (26, 27). Despite these uncertainties, the Food and Drugs Administration has recently granted accelerated approval to eteplirsen in the United States of America, recognizing that a demonstrated increase in a biologically relevant biomarker, i.e. dystrophin, however small, may reasonably be expected to benefit DMD patients (6). Hopefully, successful confirmatory trials will allow the clinical efficacy of exon skipping AONs to be established without ambiguity. Clinical trials of AONs targeting exons other than 51 are underway (e.g. NCT02310906 and NCT02500381), and new generations of AONs are being experimented at the preclinical stage (28, 29), so that exon skipping still represents a promise of future effective treatments for a large portion of DMD patients.
Stop codon readthrough compounds
About 15% of the causative mutations in DMD are single nucleotide substitutions introducing a premature termination codon (nonsense mutations) (10). This causes the ribosomial complex to stall during translation, usually resulting in nonsense-mediated decay of the transcript and absence of dystrophin (30). However, nonsense mutations may also be observed in association with BMD or intermediate phenotypes, probably due to naturally occuring alternative splicing of in-frame exons (31). Furthermore, ribosomes may be pharmacologically induced to "read through" premature stop codons, and continue downstream translation, giving rise to normal dystrophin. Aminoglycosides were demonstrated to effectively promote dystrophin expression in the mdx mouse model, which carries a nonsense mutation in exon 23 (32), but human trials were hindered by excessive toxicity (33, 34). The pharmaceutical company PTC Therapeutics has developed a small molecule compound, ataluren, which has been shown to maintain the same nonsense readthrough effect as aminoglycosides (although the exact molecular mechanism has not been completely cleared) (35) with a more favorable tolerability profile. After proof-of-concept studies in animal models (36) and phase 1 trials showing no relevant safety issues (37), ataluren was dosed in DMD in a first phase 2a study with further reassurance of safety, and encouraging results (38). The results of a larger phase 2b trial were controversial (39), as the primary endpoint (48-week change in the 6 minute walk test [6MWT]) was not achieved, and better ambulation outcomes were unexpectedly observed in a 10, 10, 20 mg/kg than 20, 20, 40 mg/kg study arm. Unfortunately, quantitative assessments of dystrophin restoration in muscle tissue from participant biopsies were hindered by technical issues in this study. However, indications of efficacy in the phase 2b study were sufficient for the European Medicines Agency to issue a conditional approval for the marketing of ataluren in European Union countries in 2014 (5), with the obligation on the company's part to conduct a phase 3 confirmatory study, the results of which (NCT01826487) have recently been released online (http://ir.ptcbio.com/releasedetail.cfm?ReleaseID=936905) although not yet published as a peer-reviewed article. While the primary endpoint of change in the 6MWT still remained elusive, pre-specified subgroup analyses and meta-analyses in conjunction with the previous phase 2b study showed a clear, although not dramatic drug effect in delaying the deterioration of ambulatory function in DMD. Currently, ataluren is prescribable to DMD patients with nonsense mutations who are older than 5 years and ambulatory in several European countries. The continued development of the nonsense readthrough strategy will hopefully provide a solid therapeutic option for a relevant subgroup of DMD patients.
"Exon snipping": mutation-specific gene editing
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems are an adaptive immune defense evolved in bacteria and archaea, which uses short RNAs for the degradation of foreign nucleic acids. In 2013, several independent groups reported the successful reprogramming of CRISPR-Cas9 to cut DNA at any site of choosing in eukaryotic cells, by coupling it with a strand of guide RNA (gRNA) with a custom sequence (40-42). This breakthrough invention has revolutionized molecular biology with its myriad potential applications. Applied to DMD, the CRISPR-Cas9 approach has been aptly named "exon snipping" (43), in analogy with the "exon skipping" obtained by AON targeting of mRNA. "Exon snipping" would primarily consist in excising an in-frame exon containing a small nonsense or frameshifting mutation, as successfully demonstrated with adeno-associated virus (AAV) delivery in the mdx murine model (44, 45), or, alternatively, in excising specific exons in order to restore the ORF in the case of large rearrangements (deletions or duplications) (46). The main drawbacks of this approach are represented by the challenges of AAV vector delivery in humans, and the fear of off-target effects. Nevertheless, the versatility and wide applicability of this technology to virtually every DMD causing mutation makes it one of the most exciting and promising novel approaches to molecular therapy of DMD.
Conclusions
The advancements of molecular treatments described above have made reaching a precise genetic diagnosis in DMD more and more important over the last few years. Currently, DMD patients with deletions bordering exon 44, 45, 51 or 53 bordering may be eligible for recruitment in one of several ongoing clinical trials of exon skipping. Patients eligible for exon 51 skipping may be treated with commercialized eteplirsen in the United States (Exondys 51®, Sarepta Therapeutics, Cambridge, MA, USA). Patients in whom MLPA (or other equivalent quantitative assays for large deletions/duplications) tests negative, should be studied with DMD gene sequencing in order to be able to provide genetic counseling to the family, and because patients with nonsense mutations may be eligible for treatment with ataluren, commercialized as Translarna® in the European Union (PTC Therapeutics, South Plainfield, NJ, USA), or within future clinical trials. While the advancement of these treatments has been painstakingly slow in the eyes of DMD patients and their families, who struggle every day against the progression of this disabling disease, there are reasons to hope that the experience gathered in designing better clinical trials, as well as an increasing number of novel drugs in the pipeline of preclinical research, will bring on a faster and more effective translation of scientific findings into benefit for patients in the upcoming years.
Acknowledgements
This work was supported by a University of Padova grant (CPDA151054 to E.P.).
References
- 1.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Fischbeck KH, Brown RH, et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 3.Bello L, Piva L, Barp A, et al. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 2012;79:159–162. doi: 10.1212/WNL.0b013e31825f04ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricotti V, Muntoni F, Voit T. Challenges of clinical trial design for DMD. Neuromuscul Disord. 2015;25:932–935. doi: 10.1016/j.nmd.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Haas M, Vlcek V, Balabanov P, et al. European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscul Disord NMD. 2015;25:5–13. doi: 10.1016/j.nmd.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Aartsma-Rus A, Krieg AM. FDA approves eteplirsen for duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bello L, Hoffman EP, Pegoraro E. Muscular Dystrophy: Causes and Management. 2013. Dystrophinopathies; pp. 67–96. [Google Scholar]
- 8.Bushby K, Finkel R, Birnkrant DJ, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 9.Falzarano MS, Scotton C, Passarelli C, et al. Duchenne muscular dystrophy: from diagnosis to therapy. molecules. 2015;20:18168–18184. doi: 10.3390/molecules201018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aartsma-Rus A, Deutekom JCT, Fokkema IF, et al. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 11.Janssen B, Hartmann C, Scholz V, et al. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005;6:29–35. doi: 10.1007/s10048-004-0204-1. [DOI] [PubMed] [Google Scholar]
- 12.Monaco AP, Bertelson CJ, Liechti-Gallati S, et al. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 13.Saleh AF, Arzumanov AA, Gait MJ. Overview of alternative oligonucleotide chemistries for exon skipping. Methods Mol Biol Clifton NJ. 2012;867:365–378. doi: 10.1007/978-1-61779-767-5_23. [DOI] [PubMed] [Google Scholar]
- 14.Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 15.Kinali M, Arechavala-Gomeza V, Feng L, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goemans NM, Tulinius M, Akker JT, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 17.Cirak S, Arechavala-Gomeza V, Guglieri M, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet Lond Engl. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voit T, Topaloglu H, Straub V, et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q-L, Cirak S, Partridge T. What Can We Learn From Clinical Trials of Exon Skipping for DMD? Mol Ther Nucleic Acids. 2014;3:e152–e152. doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman EP, McNally EM. Exon-skipping therapy: a roadblock, detour, or bump in the road? Sci Transl Med. 2014;6:230fs14–230fs14. doi: 10.1126/scitranslmed.3008873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anthony K, Arechavala-Gomeza V, Taylor LE, et al. Dystrophin quantification: Biological and translational research implications. Neurology. 2014;83:2062–2069. doi: 10.1212/WNL.0000000000001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricotti V, Muntoni F, Voit T. Challenges of clinical trial design for DMD. Neuromuscul Disord NMD. 2015;25:932–935. doi: 10.1016/j.nmd.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Godfrey C, Muses S, McClorey G, et al. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. Hum Mol Genet. 2015;24:4225–4237. doi: 10.1093/hmg/ddv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendell JR, Rodino-Klapac LR, Sahenk Z, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 25.Mendell JR, Goemans N, Lowes LP, et al. Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unger EF, Califf RM. Regarding eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2017;81:162–164. doi: 10.1002/ana.24842. [DOI] [PubMed] [Google Scholar]
- 27.Mendell JR. Eteplirsen improves function and partially restores dystrophin. Ann Neurol. 2016 [Google Scholar]
- 28.Gao X, Zhao J, Han G, et al. Effective dystrophin restoration by a novel muscle-homing peptide-morpholino conjugate in dystrophin-deficient mdx mice. Mol Ther J Am Soc Gene Ther. 2014;22:1333–1341. doi: 10.1038/mt.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goyenvalle A, Leumann C, Garcia L. Therapeutic potential of tricyclo- DNA antisense oligonucleotides. J Neuromuscul Dis. 2016;3:157–167. doi: 10.3233/JND-160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nigro V, Politano L, Nigro G, et al. Detection of a nonsense mutation in the dystrophin gene by multiple SSCP. Hum Mol Genet. 1992;1:517–520. doi: 10.1093/hmg/1.7.517. [DOI] [PubMed] [Google Scholar]
- 31.Flanigan KM, Dunn DM, Niederhausern A, et al. Nonsense mutation-associated Becker muscular dystrophy: interplay between exon definition and splicing regulatory elements within the DMD gene. Hum Mutat. 2011;32:299–308. doi: 10.1002/humu.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barton-Davis ER, Cordier L, Shoturma DI, et al. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politano L, Nigro G, Nigro V, et al. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol Myopathies Cardiomyopathies Off J Mediterr Soc Myol. 2003;22:15–21. [PubMed] [Google Scholar]
- 34.Malik V, Rodino-Klapac LR, Viollet L, et al. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010;67:771–780. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 35.Sheridan C. Doubts raised over "read-through" Duchenne drug mechanism. Nat Biotechnol. 2013;31:771–773. doi: 10.1038/nbt0913-771. [DOI] [PubMed] [Google Scholar]
- 36.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 37.Hirawat S, Welch EM, Elfring GL, et al. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol. 2007;47:430–444. doi: 10.1177/0091270006297140. [DOI] [PubMed] [Google Scholar]
- 38.Finkel RS, Flanigan KM, Wong B, et al. Phase 2a study of atalurenmediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PloS One. 2013;8:e81302–e81302. doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014;50:477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNAguided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kemaladewi DU, Cohn RD. Exon Snipping in Duchenne muscular dystrophy. Trends Mol Med. 2016;22:187–189. doi: 10.1016/j.molmed.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ousterout DG, Kabadi AM, Thakore PI, et al. Multiplex CRISPR/ Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244–6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]