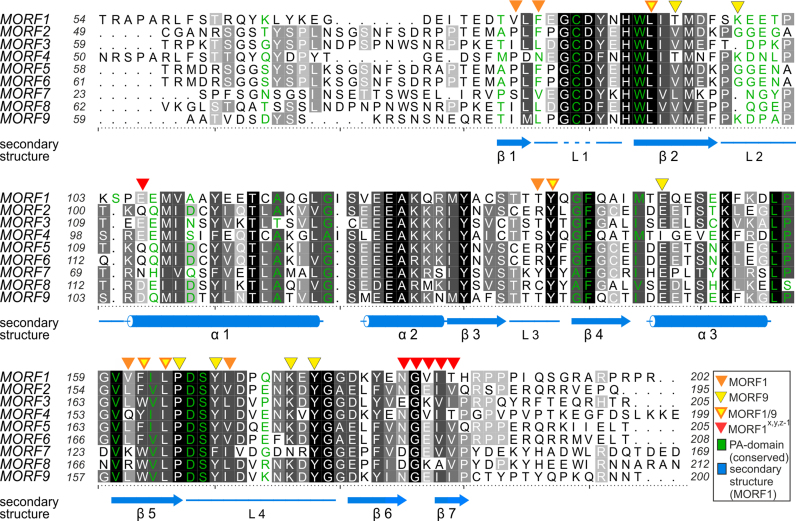
Figure 3.
Alignment of MORF domains of Arabidopsis thaliana. The alignment was prepared by Chimera employing Clustal Omega (40) and shaded with ALSCRIPT (47). Proteins are identified on the left of the aligned sequences. Higher conservation is indicated by a darker background. Numbering refers to the respective proteins. Below the alignment, secondary structure elements (α – α-helix, β – β-sheet, L – loop) of MORF179–190 are shown in marine blue and numbered, a dashed line indicates the modeled loop of residues G84-D86. Orange triangles above the alignment indicate residues of MORF179–190, which are forming the main hydrophobic dimerization interface. Yellow triangles indicate the MORF986–186 dimerization interface. Yellow triangles with orange frame mark positions of residues which both contribute to the dimerization interface of MORF179–190 and MORF986–186. Red triangles above the alignment mark residues of MORF179–190 that form a secondary interface as observed in two different crystal lattices. Green - Residue identities between MORF179–190 and the PA domain of human proprotein convertase subtilisin/kexin type 9 (sequence of PDB ID: 2W2N).