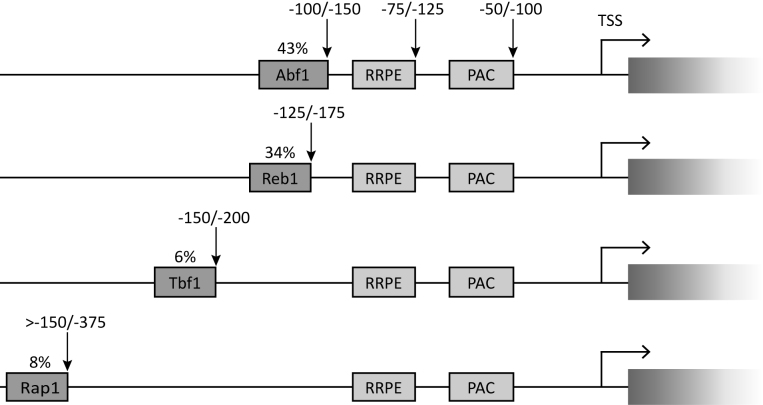
Figure 1.
Conserved sequence elements in Saccharomyces Ribi gene promoters. Graphical representations of promoter architecture of Ribi genes as derived from in silico analysis. The ∼20% of Ribi genes which did not display any conserved GRF binding site is not represented in the Figure. Indicated above each GRF box is the percentage of Ribi gene promoters in which the motif occurs. Vertical arrows pointing to the downstream end of the boxes are to indicate their range of distances (in base pairs, above the arrows) with respect to the TSS. The consensus sequences considered for the different elements reported are: PAC, GCGATGAGMT; RRPE, TGAAAAWTTTY; Abf1, RTCAYNNNN(N)ACGR; Reb1, RTTACCCK; Tbf1, ARCCCTAA; Rap1, WACAYCCRTACATY (M, A or C; W, A or T; R, A or G; Y, C or T; K, G or T; N, any nucleotide). These consensus sequences are based on the following references: PAC and RRPE (1); Abf1 (10,12,55); Reb1 (10,55); Tbf1 (10,12,18); Rap1 (56).