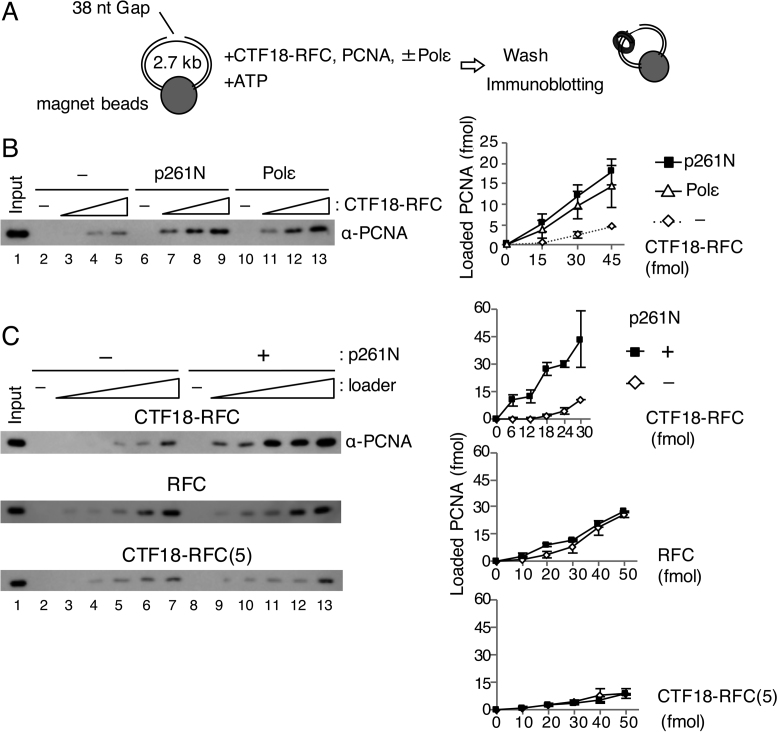
Figure 1.
PCNA loading by CTF18-RFC in the presence of Polε. (A) Schematic diagram of the PCNA loading assay with a gapped DNA attached to magnetic beads. A linearised 2.7 kb plasmid DNA harbouring a 38 nt gap was used. After incubation of the DNA beads with purified proteins and ATP, the loaded PCNA was recovered from the bead-bound fraction. (B) PCNA loading by 15–45 fmol of CTF18-RFC with either 100 fmol of p261N (lanes 6–9) or 100 fmol of Polε (lanes 10–13), or with neither (lanes 2–5). The 10 μl reaction mixture contained 30 mM NaCl, 40 mM creatine phosphate and 25 ng/μl creatine-phosphate kinase. Input control (17 fmol of trimeric PCNA) (lane 1) and 50% of bound samples (lanes 2–13) were analysed by immunoblotting with anti-PCNA antibody. The loaded PCNA was quantified and graphed with mean ± S.E. of two experimental replicates (right). (C) PCNA loading by 6–30 fmol of CTF18-RFC (top), 10–50 fmol of RFC (middle) or 10–50 fmol of CTF18-RFC(5) (bottom) in the same reaction mixture with (+) or without (−) 200 fmol of p261N. Input (lane 1 of each) used 17 fmol (top and middle) or 12 fmol (bottom) PCNA. The bound fractions (50%) were analysed for loaded PCNA (lanes 2–13), which was quantified and graphed as indicated at the right with mean ± S.E. of two experimental replicates.