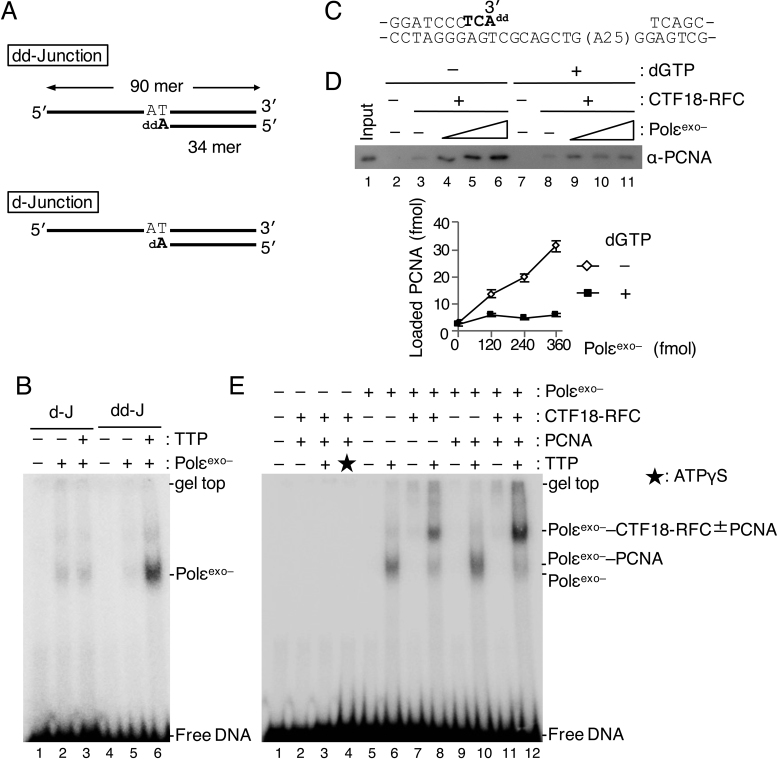
Figure 6.
Analyses of PCNA loading in the presence of Polε in synthesis mode. (A) Two 3΄ primer–template junction substrates with ddAMP (“dd-Junction”) or dAMP (“d-Junction”) at their 3΄ primer ends. (B) 60 fmol of Polεexo– was mixed with 25 fmol of d-Junction (“d-J”; lanes 2, 3) or dd-Junction (“dd-J”; lanes 5, 6) at 60 mM NaCl in the presence (+) or absence (−) of 100 μM TTP, and binding was analysed by EMSA after glutaraldehyde fixation. Lanes 1 and 4 were controls without Polεexo–. Bands produced by binding of Polεexo– to DNA are indicated. (C) To study PCNA loading in the presence of Polεexo– in synthesis mode, a gapped DNA with ddAMP at the 3΄ primer end was prepared. The sequence shows a 51 bp region with the 35 nt gap on the substrate DNA. The nucleotides shown in bold represent sequence extension to prepare the 3΄ primer end with ddAMP. (D) Comparison of PCNA loading with the gapped-DNA beads with Polεexo– in non-synthesising (–dGTP) and synthesising (+dGTP) modes. The indicated DNA beads (15 ng) were incubated with 100 fmol of CTF18-RFC and 6.2 pmol of PCNA in the presence of 0, 120, 240 and 360 fmol of Polεexo– in a 10 μl reaction mixture at 60 mM NaCl. dGTP (100 μM) was added in lanes 7–11. Input control of 12 fmol of PCNA (lane 1) and 100% bound fractions (lanes 2–11) were applied to immunoblotting with anti-PCNA antibody. Lanes 2 and 7 were the negative controls without CTF18-RFC and Polεexo–. Bound PCNA was quantified and graphed below with mean ± S.E. of two experimental replicates. (E) Assembly of 60 fmol each of CTF18-RFC and Polεexo– and 500 fmol of PCNA to 25 fmol of dd-Junction substrate was analysed by EMSA as above (B) using indicated combinations of proteins at 60 mM NaCl. Additions of 100 μM TTP and 250 μM ATPγS are indicated by ‘+’ and an asterisk (lane 3), respectively. DNA bands shifted at positions by added proteins are indicated at the right.