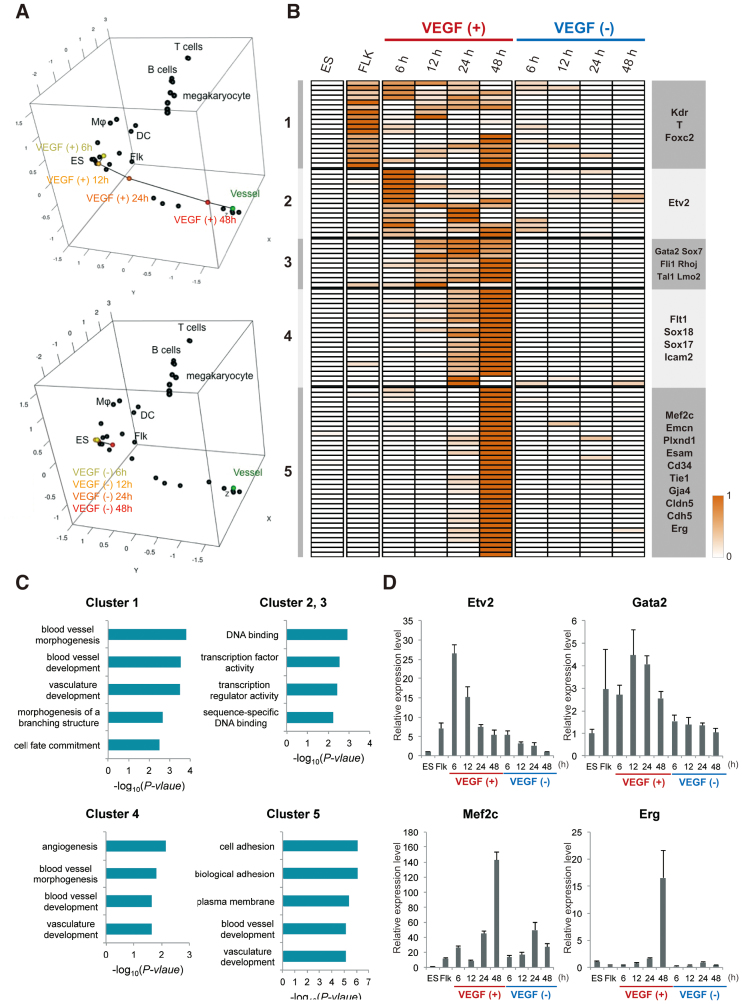
Figure 2.
Global and dynamic transcriptome analysis on the EC differentiation. (A) Three-dimensional (3D) MDS plots. The cells in the presence or absence of VEGF and the normal mouse tissues were mapped to the 3D space based on the expression profiles of the 79 EC-specific genes by using classical MDS. Black dots represent each cell line. Connected black line indicates gradual differentiation to mature ECs. Upper and lower 3D MDS indicate in the presence or absence of VEGF, respectively. DC and Flk into the box represent dendritic cells and Flk-sorted mesoderm cells, respectively. (B) Heat map representation of the VEGF responsive genes. Genes were classified based on the VEGF treated time points using the algorithm ‘HOPACH’. (C) Functional annotations for each cluster are shown. The P-values of each category analysed from DAVID are shown in the bar graphs. (D) Validation of the representative genes in (B) was conducted by the qPCR. The graph value indicates the expression levels relative to in ES cells. Data are shown as the mean and the standard deviation from triplicates.