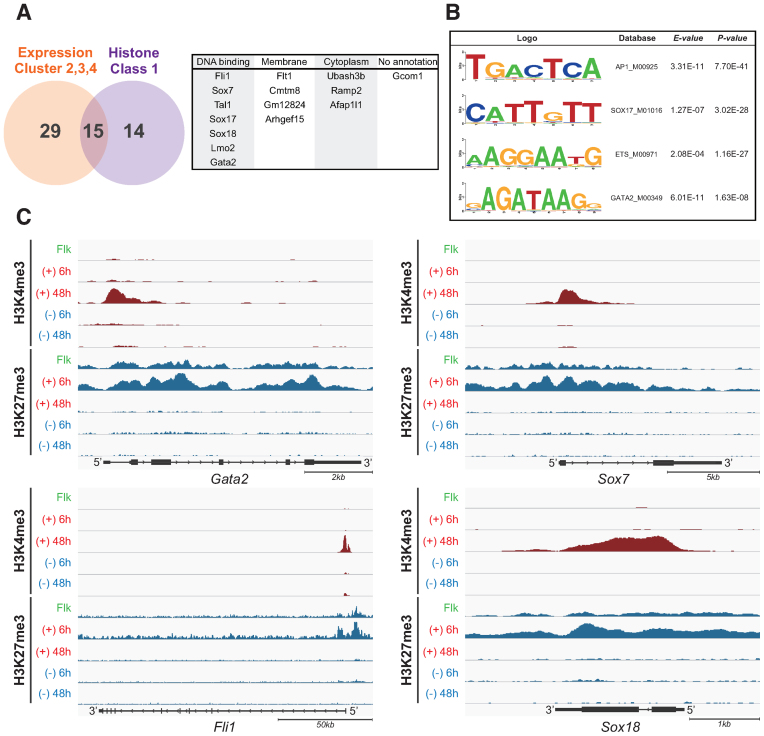
Figure 4.
Identification of the master transcription factor for EC differentiation. (A) Venn diagram depicting the overlap of expression cluster 2,3 and 4 in Figure 2B and histone class 1 in Figure 3C. The numbers indicate the overlapped or unique genes count. The common 15 genes are classified by GO term (right panel). (B) Determination of enriched transcription factor motifs calculated from the enhancer regions of HUVECs database. The MODIC method (32) was used for identification of enriched sequences and the size of the character reflects the degree of enrichment. E-value and P-value indicate the probability that de novo enriched sequences obtained from FAIRE-seq are matched to the shown ‘Web logo’ and known consensus motifs by chance, respectively. (C) UCSC mm9 genome browser view around Gata2, Fli1, Sox7 and Sox18 genes. ChIP-seq tags were calculated by MACS and are shown in IGV. H3K4me3 peaks were shown in red, and H3K27me3 were in blue.