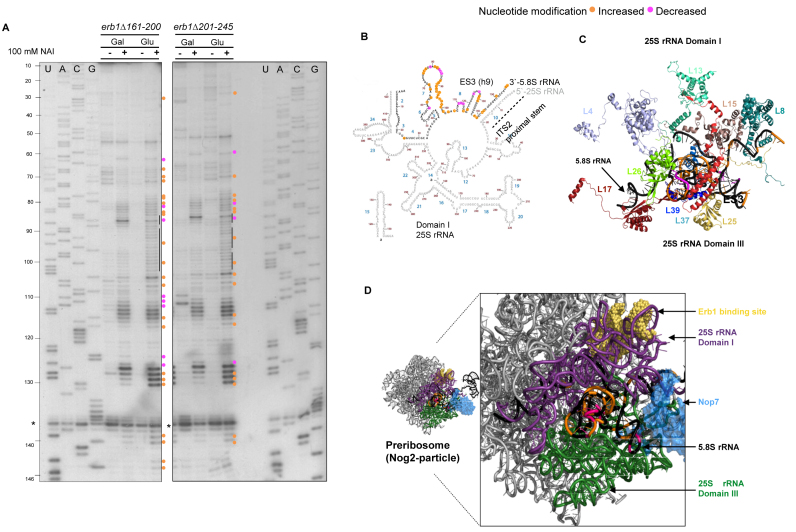
Figure 5.
5.8S rRNA is not folded properly in the erb1Δ161–200 and erb1Δ201–245 mutants. (A) In vivo SHAPE assay of 5.8S rRNA structure in GAL-ERB1 + erb1 mutants grown in galactose, or grown in galactose and shifted to glucose. Nucleotides with increased or decreased modification are indicated with orange and pink circles, respectively. RNA isolated from cells treated with DMSO alone were used as controls (indicated by –). The sequencing ladders are shown and nucleotide positions are indicated (U, A, C, G) for 5.8S rRNA. The common stop (*) in pre-rRNA was used to assess loading. (B) Locations of nucleotides modified in the erb1 mutants are displayed on the 5.8S rRNA/domain I of 25S rRNA structure (www.ribovision.org). Nucleotide numbers (brown) and helices (blue) in 5.8S rRNA are marked. (C) PyMOL representation of modified residues in 5.8S rRNA with the r-proteins that bind it (PDB ID: 4V88). (D) The 5.8S rRNA is sandwiched between 25S rRNA domains I (purple) and III (green) (PDB ID: 3JCT) (28). The structure of Nop7 protein (blue) and the rRNA binding site of Erb1 (yellow) are also shown.