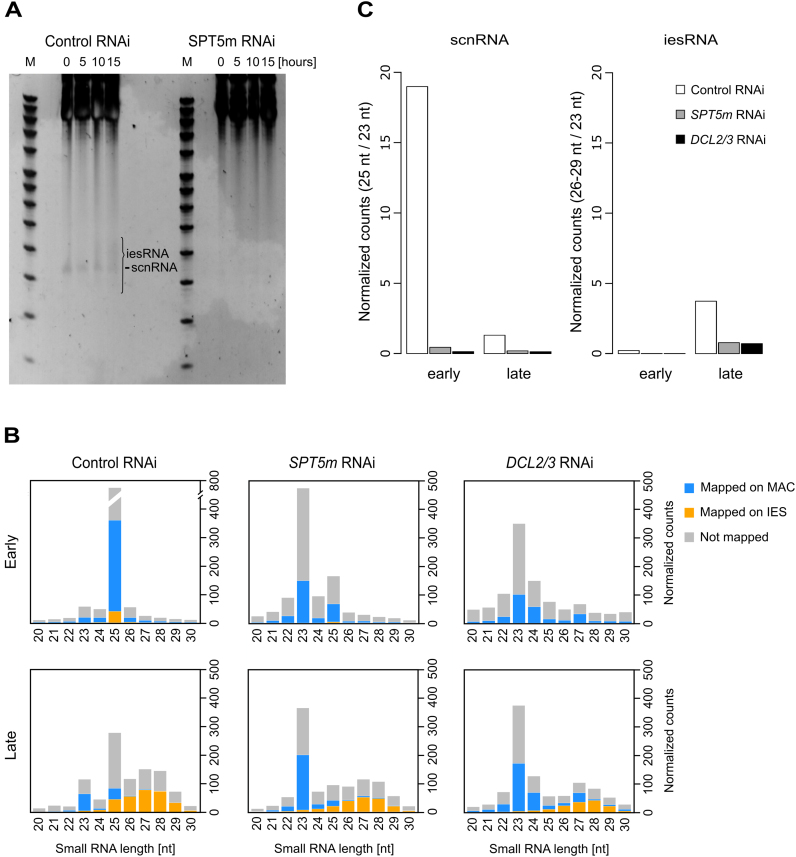
Figure 3.
Analysis of sRNA populations in SPT5m-RNAi cells. (A) Gel electrophoresis of sRNA from SPT5m-silenced cells and control silencing. Total RNA samples corresponding to the T0, T5, T10 and T15 time-points were run on a denaturing 15% polyacrylamide-urea gel. After electrophoresis the gel was stained with SYBR Gold (Invitrogen). M: DNA Low Molecular Weight Marker (USB). The ∼25 nt signal that corresponds to the fraction of scnRNAs (29) is labeled. In the control, at the T15 time-point, additional bands corresponding to 26–30 nt iesRNAs are present (indicated by a bracket). In SPT5m-silenced samples neither scnRNAs nor iesRNAs can be seen. (B) Small RNA libraries corresponding to early and late development time-points were sequenced and mapped to the reference genomes (Paramecium tetraurelia MAC reference genome and MAC+IES reference genome). Results obtained for a control culture (cells silenced for ND7 gene expression) are shown in the left hand panel, SPT5m-silencing in the middle panel and DCL2/3-silencing (from (30)) in the right hand panel. Stacked bar plots show the normalized number of sRNA reads that match the MAC genome (blue), annotated IESs (yellow) or were not mapped (gray). (C) Read counts obtained during sequencing of small RNA libraries corresponding to early and late development time-points were normalized. Bar plots show the relative amounts of sRNA reads for control (white), SPT5-RNAi (gray) and DCL2/3-RNAi (black, for details of see (30)).The left hand panel corresponds to scnRNA and shows the number of 25 nt reads divided by the number of 23 nt siRNA reads for each sample. Right hand panel shows iesRNA reads (26–29 nt) in relation to 23 nt reads. Since iesRNAs are supposed to be produced by Dcl5 from excised IESs (30), their appearance in the late sample could be reduced owing to reduced IES excision in SPT5 and DCL2/3-RNAi.