Figure 6.
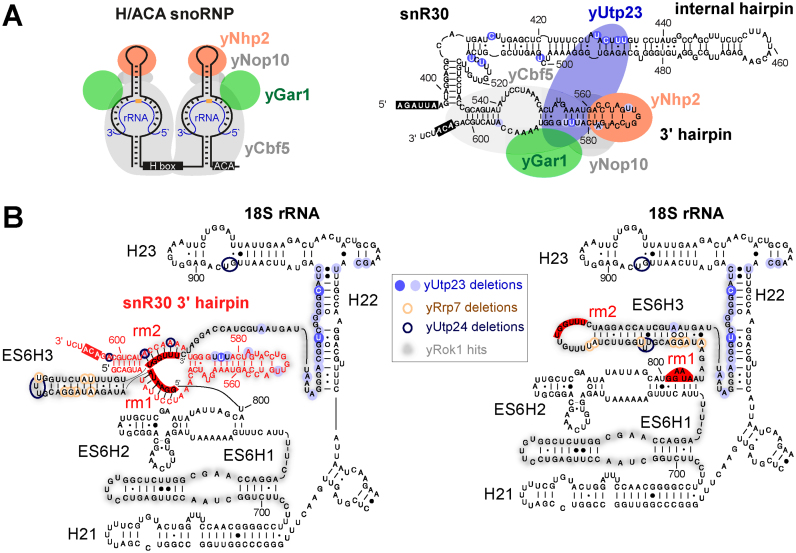
Yeast yUtp23 is central to the integration of ES6 binding factors and snR30 release. (A) Left panel: Cartoon of a typical modification H/ACA box yeast snoRNP depicting the positions of the core H/ACA snoRNP proteins yCbf5, yGar1, yNop10 and yNhp2. H and ACA boxes are shown in black. Orange circle: target nucleotide for pseudouridylation in the rRNA (blue). Right panel: Cartoon of the 3΄ terminal portion of yeast snR30 with the predicted positions of yUtp23 and core H/ACA snoRNP proteins. yUtp23 microdeletion sites in the internal hairpin and 3΄ hairpin are represented as blue circles, shades represent peak height as in Figure 2C. The H and ACA boxes are shown in black. (B) Predicted structure of the yeast 18S ES6 region during snR30 base-pairing (left panel) and in the mature rRNA after snR30 release (right panel). Precise crosslinking sites of yUtp23 (blue circles, shades indicate peak height as in Figures 1 and 2), yUtp24 (dark blue) and yRrp7 (brown), and yRok1 crosslinking regions (shaded in grey) on the snR30 3΄ hairpin (red) and 18S rRNA sequences (black). Binding sites for snR30 in the ES6 region (rm1 and rm2) and the ACA box are highlighted in red.