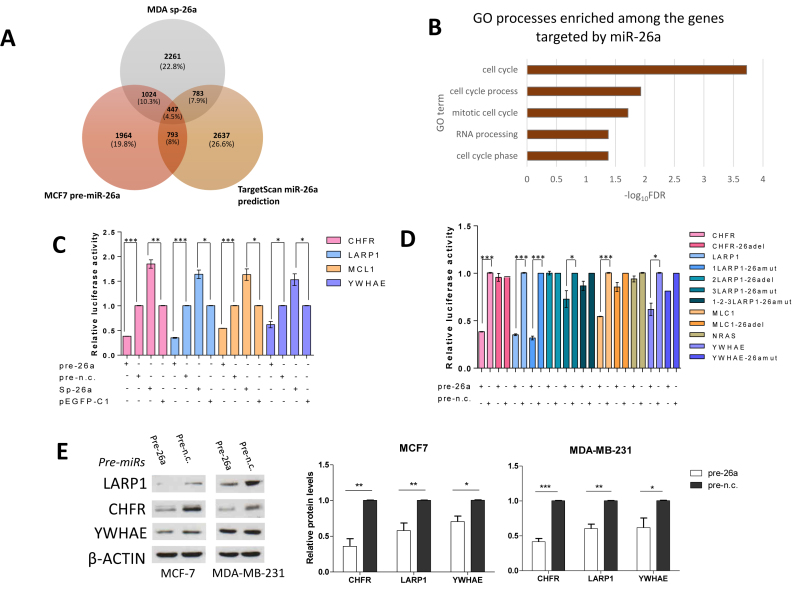
Figure 3.
Three mitosis-related genes are novel miR-26a targets. (A) Venn diagram representing the number of possible miR-26a targets identified as an area of overlap between transcripts downregulated in MCF cells over-expressing miR-26a, MDA-MB-231 cells with reduced miR-26a level and TargetScan-predicted miR-26a target genes. (B) Chart showing the top five significantly enriched GO terms found with DAVID bioinformatic tool amongst the possible miR-26a targets identified as described in panel A. (C and D) MiR-26a targets CHFR, LARP1, MCL1 and YWHAE by directly interacting with their relative 3΄UTRs. Relative luciferase activity levels were measured after 24 h from co-transfection of MCF-7 cells with the indicated 3΄UTR-luciferase reporter constructs either with miR-26a or miR-n.c. precurors (100 nM) or with mi-26a-sponge construct or pEGFP-C1 parental control (150 ng). Panel D shows luciferase activity levels in cells expressing mutant 3΄UTRs which impair miR-26a binding. Data are mean of three independent experiments (each of them performed in triplicate) ± S.E.M. (*P < 0.05,**P ≤ 0.006,***P ≤ 0.0005). (E) Western blots showing CHFR, LARP1 and YWHAE levels after transfection for 9 days of the two cell lines with the indicated miRNA precursors (5 nM) and relative densitometric quantifications. β-Actin was used as a loading control. Fold changes in protein expression levels normalised for β-actin using ImageJ software are shown underneath each relative protein plot (*P < 0.01, **P ≤ 0.001, ***P ≤ 0.0001).