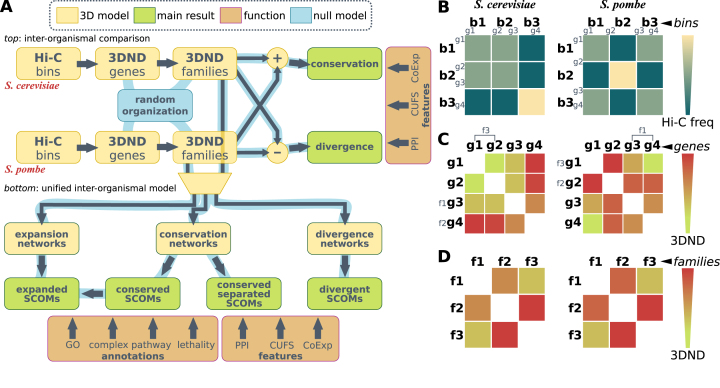
Figure 1.
Research plan. (A) Flow chart depicting the analysis pipeline. Component types (model, result, function, null model) are denoted by color. Top: representing 3D network distances (3DND) between orthologous families in each organism (data in bold and coordinates in regular font); analysis of conservation and divergence of organization vs. functional features. Random genomic organizations are generated and passed down the pipeline to obtain an empirical p-value for each result. Bottom: a unified network analysis of conservation and divergence of gene families; detection of spatially co-evolving ortholgous modules (SCOMs) and their analysis vs. functional annotations and features. Random genomic organizations are passed down the pipeline as well. (B) An illustration of the preparation of an inter-organismal model, including the first 3 steps shown in panel (A) (for network and SCOM illustrations see Figures 2–5). Normalized Hi-C frequency/probability matrices containing three bins are shown for S. cerevisiae (left) and S. pombe (right). Chromosome conformation is different in each species, and gene distribution (four genes marked) is different. (C) Continuation of the procedure in panel (B). Hi-C network distance matrices after transformation to gene coordinates are shown for the two organisms. Three orthologous families (with different structure/location) are marked. (D) Continuation of the procedure in panel (C). Distance matrices after transformation to family coordinates are shown for the two organisms. Despite differences in conformation, gene location and family structure, the resulting distances are conserved. These distances are utilized to generate the various networks, such as the conservation network.