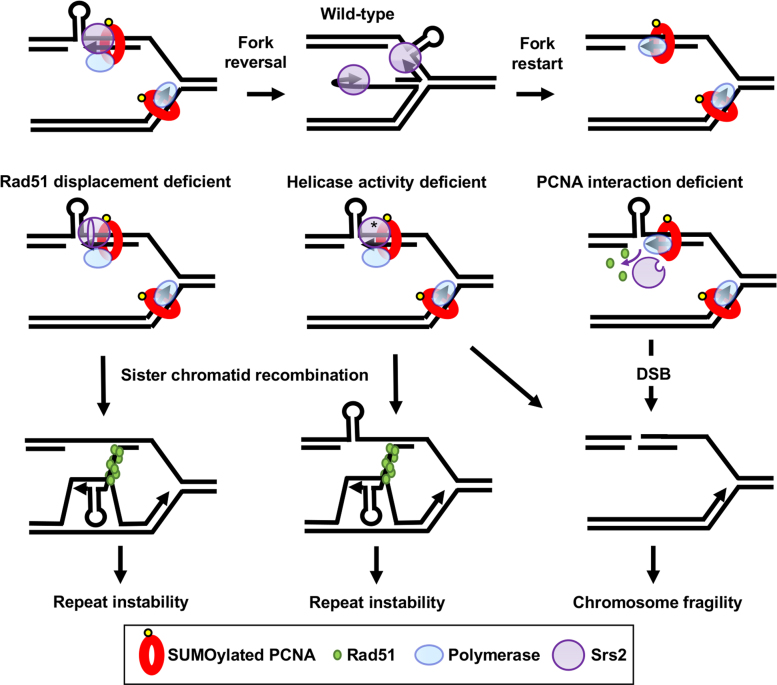
Figure 5.
A model to explain how the different domains of the Srs2 protein are involved in repeat instability, chromosomal fragility and JM formation. Top row: in the wild-type situation Srs2 is brought to the fork by PCNA interaction and unwinds hairpins formed by triplet repeats. For simplicity, only a lagging strand template hairpin is shown, though we note that leading strand template or nascent strand hairpins are also possible. Transient fork reversal may facilitate unwinding, or reversal may occur when unwinding fails, forming some JMs in wild-type cells (reversed fork JMs, Rad51 independent). Homologous recombination is discouraged by Srs2-dependent Rad51 displacement, but may occasionally happen. Right: in the absence of interaction with PCNA, Srs2 is not recruited to the fork to unwind template hairpin structures, leading to fork breakage. Srs2 may still displace Rad51, preventing recombination-dependent instability. Center: when Srs2 ATPase activity is absent (*) hairpins will not be unwound, leading to more fork breakdown, and Rad51 is not displaced, leading to increased recombination intermediates. In this case, JMs are increased, consisting of both reversed forks and recombination intermediates. Left: Srs2 missing the Rad51 interaction domain is brought to the fork by PCNA interaction and can unwind secondary structures to prevent fork breakdown, but cannot displace Rad51p. Increased homologous recombination occurs, leading to repeat instability.