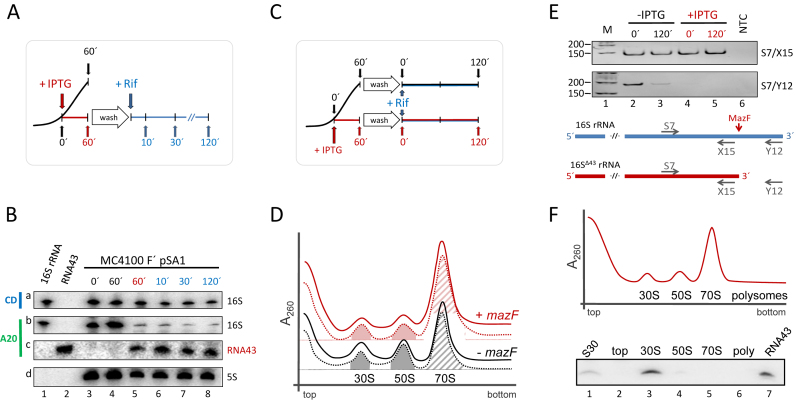
Figure 2.
RNA43, 16SΔ43 rRNA and 70SΔ43 ribosomes are stable during stress conditions. (A) Schematic depiction of the experimental approach to assess the stability of RNA43 and 16SΔ43 rRNA in vivo. A schematic growth curve in the absence of IPTG is shown in black. At OD600 of 0.3 the culture was divided and IPTG was added to one half to induce mazF expression (growth is blocked as indicated in red). 60 minutes thereafter, cells were washed and resuspended in fresh medium comprising rifampicin (in blue). (B) Samples withdrawn at the time points indicated were subjected to northern blot analysis with probes specific for the central domain of the 16S rRNA (CD, panel a), the 3΄-terminus of the 16S rRNA (A20; panel b) and the RNA43 (A20; panel c). 5S rRNA was used as internal standard for quantification (panel d). In vitro transcribed 16S rRNA (lane 1) and RNA43 (lane 2) served as size markers. The experiment was performed in triplicate and one representative autoradiograph is shown. (C) Schematic of the experimental approach to assess the stability of 70SΔ43 ribosomes in vivo as described in A. (D) S30 extracts were prepared before (dotted lines) and 120 min after addition of rifampicin (solid lines) to untreated cells (black lines) or 60 min after induction of mazF expression (red lines) and subjected to sucrose density gradient analysis. Peaks representing 30S and 50S subunits and 70S ribosomes are indicated. The peak areas of the 30S and 50S subunits (filled areas) and 70S monosomes (hatched area) that were quantified to determine the subunits/monosome ratios are indicated. (E) To monitor MazF-mediated processing of the 16S rRNA, RNA was isolated from the fractions comprising the 70S monosomes. RT-PCR analysis using primers S7/X15 (upper panel), specific for both intact 16S rRNA and 16SΔ43 rRNA, and primers S7/Y12 (lower panel), yielding a product only with uncleaved 16S rRNA. NTC: no template control. Below, the binding sites of the primers are given schematically. (F) Ribosome sedimentation profiles of cell extracts 30 min after induction of mazF expression. Total RNA was purified from the indicated fractions (top, 30S, 50S, 70S and polysomes), respectively, and tested for the presence of RNA43 by northern blotting. Total RNA purified from the S30 extract withdrawn 30 min upon induction of mazF expression (lane 1), and in vitro transcribed RNA43 (lane 7) served as controls.