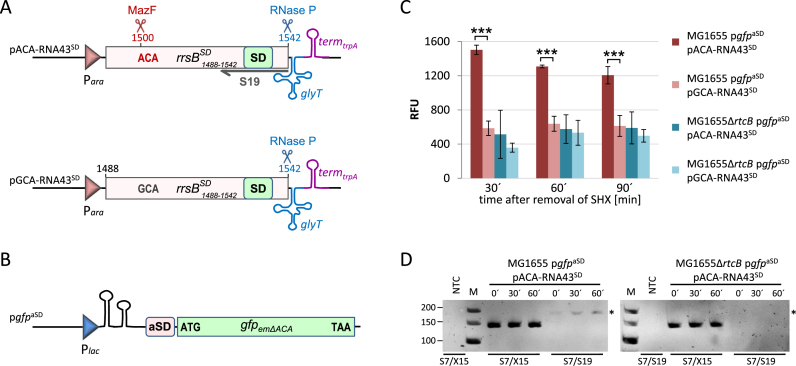
Figure 6.
RtcB restores the ability of the ribosomes to translate canonical mRNAs in vivo. (A) Schematic depiction of the construct encoded by plasmid pACA-RNA43SD used to ectopically express the rrsBSD fragment that can be processed by MazF at the ACA-site to generate the mature RNA43SD. Below, the control construct comprising the rrsBSD fragment without the ACA-cleavage site is shown (pACA-RNA43SD), which cannot be processed by MazF. The binding site of the primer S19 used for RT-PCR analysis shown in D is indicated. See text for details. (B) Scheme showing the corresponding gfpaSD reporter gene of plasmid pgfpaSD, which does not harbor ACA-sites not to interfere with mRNA levels due to activation of MazF during stress treatment (Oron-Gottesman et al., submitted). See text for details. (C) The relative fluorescence units (RFU, normalized to OD600) were measured for strains MG1655 and the isogenic rtcB-deletion strain harboring the gfpaSD reporter gene and either plasmid pACA-RNA43SD or plasmid pGCA-RNA43SD at the indicated time points before (0 min) and during (30 and 60 min) recovery from SHX treatment. The experiment was performed in triplicate; error bars indicate the standard deviation from the mean (***P < 0.001). (D) RT-PCR analysis to monitor the formation of the 16SSD rRNA by RtcB-mediated ligation during recovery from SHX-treatment in strains MG1655 and MG1655ΔrtcB. Primer pair S7/S19 specifically amplifies a 182 nts long product only after ligation of the 16SΔ43 rRNA and the ectopically expressed RNA43SD (indicated by an asterisk). Primers S7/X15 served as internal control. NTC: no template control. The length of the marker bands in nts is given to the left.