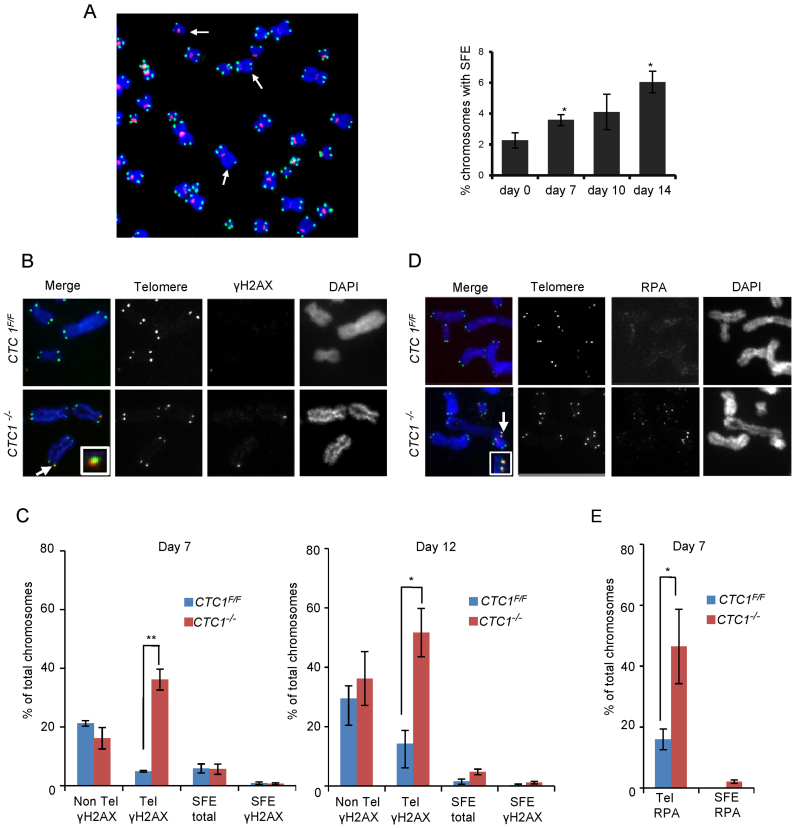
Figure 2.
CTC1 disruption causes telomeric DNA damage signaling at telomeres that retain telomeric sequence. CTC1 conditional cells were grown with/without tamoxifen for the indicated times. (A) Left: Metaphase spread from cells grown with tamoxifen for 10 days. White arrows indicate partial telomere loss or full telomere loss (signal free ends, SFE). Chromosomes were hybridized with (C3TA2)3 telomere probe (green) and centromere probe (red) and stained with DAPI (blue). Right: Quantification of chromosomes showing one or more SFEs at the indicated time points, n = 3 independent experiments, mean ± S.E.M, ≥2000 chromosomes analyzed per time point. (B–E) Metaphase spreads showing γH2AX or RPA localization. Telomeres detected by FISH (green), γH2AX or RPA by immunostaining (red), chromosomes were counterstained with DAPI (blue). n = 3 independent experiments, mean ± S.E.M. ≥300 chromosomes scored per time point. (B) Images showing γH2AX staining at day 12. Arrows indicate co-localization, inserts show enlargement of same region. (C) Quantification of chromosomes with γH2AX foci on chromosome arms (Non-Tel), at one or more telomeres with detectable telomeric DNA (Tel) or at signal free ends (SFE). (D) Images showing RPA staining at day 7. (E) Quantification of chromosomes with RPA foci at telomeres.