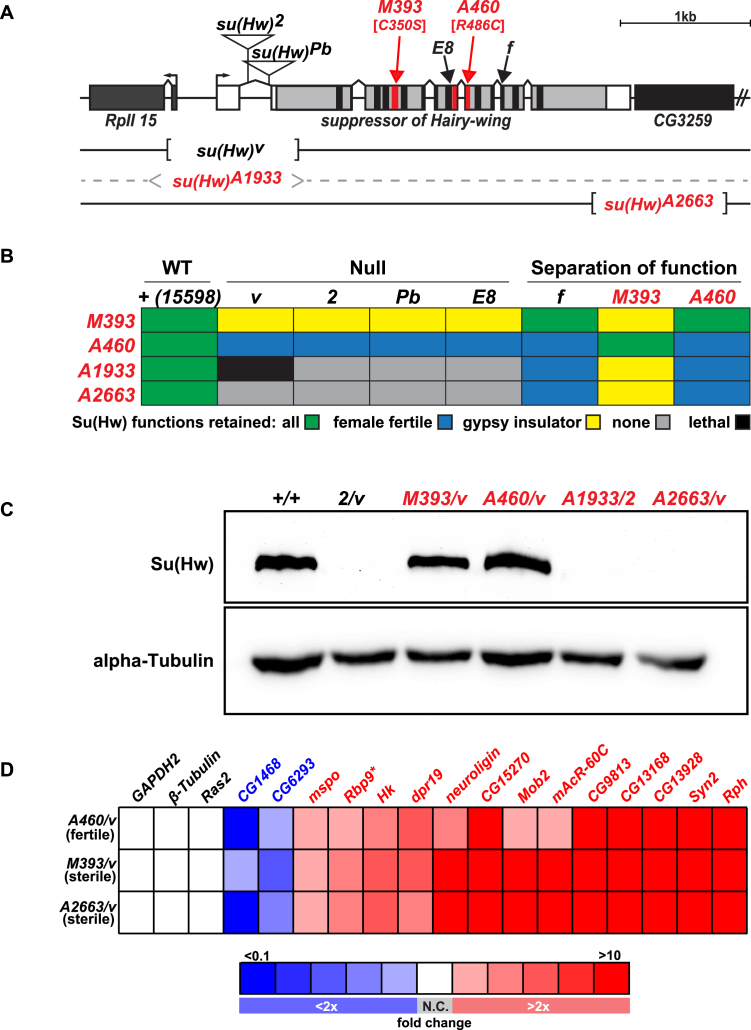
Figure 2.
Properties of su(Hw) alleles generated during EMS mutagenic screen. (A) Shown is a diagram of the su(Hw) locus, including su(Hw) and the 5΄ RPII15 and 3΄ CG3259 genes (rectangles). The 5΄ and 3΄ UTRs of the su(Hw) gene are shown in white and the coding region in gray, with the locations of the ZFs shown in black or red. Lesions associated with mutant alleles are depicted, including the insertions in su(Hw)2 and su(Hw)Pb, the ZF point mutations in su(Hw)M393 [C350S], su(Hw)E8, su(Hw)A460 [R486C] and su(Hw)f, and the location of the deletions in su(Hw)v, su(Hw)A1933 and su(Hw)A2663. (B) Complementation data obtained from crosses between extant and new su(Hw) alleles, including both null and separation-of-function mutants. Trans-heterozygotes showed the following phenotypes: (i) had all Su(Hw) functions (green), (ii) had female fertility only (blue), (iii) had gypsy-insulator function only (yellow), (iv) had no Su(Hw) function (gray) or (v) was adult lethal (black). The su(Hw)+ stock was the parental stock used in the EMS screen that carried the marked P[y+, w+]15598 third chromosome. (C) Western blot of protein extracts obtained from su(Hw)+/+ and su(Hw)−/-ovaries probed with antibodies against Su(Hw) and alpha-Tubulin (loading control). (D) Heat map of qRT-PCR analyses of gene expression changes of Su(Hw) target genes in RNA isolated from the su(Hw)A460/v (fertile), su(Hw)M393/v (sterile) and su(Hw)A2663/v (sterile) relative to the su(Hw)+/+ parental line (15 598). Genes studied are listed above the table, with three non-target genes (black) and fifteen target genes analyzed, including two upregulated (blue) and thirteen downregulated (red) genes. Boxes represent less than 2-fold (white, no change: N.C.) or 2–4-fold, 4–6-fold, 6–8-fold, 8–10-fold and greater than 10-fold changes, denoted as darkening shades of red and blue for increased and decreased, respectively. Data represent the average fold change of three biological replicates.