Figure 6.
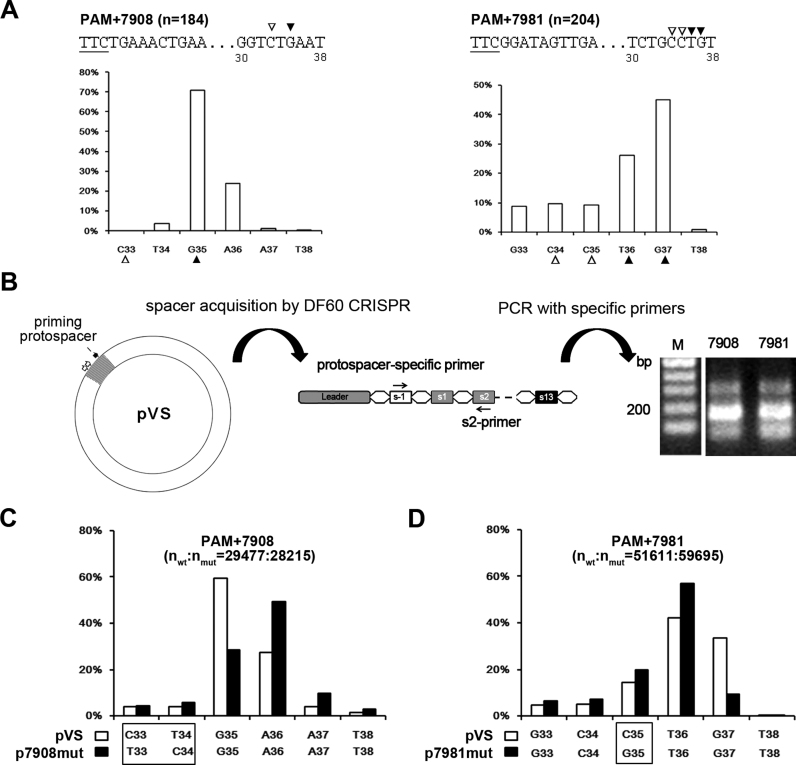
Protospacer mutations at the 3΄-end could alter the final spacer size. (A) Variation of the 3΄-end of spacers from groups PAM+7908 and PAM+7981. The horizontal and vertical coordinates, respectively, represent the PAM-distal nucleotides (33–38 bp downstream of the PAM) and the ratio of the spacers terminating at each position. The third nucleotide (indicated by an empty triangle) at the prevalent 3΄-end (indicated by a solid triangle) tended to be a cytosine. The PAM (underlined) and its following sequences are shown for each group. Position numbers shown beneath the sequence are relative to the PAM. (B) An assay to detect the acquisition of the PAM+7908 and PAM+7981 spacers from pVS which carries an HHPV-2 fragment (in gray). The viral fragment includes sequences (two empty arrows) from which PAM+7908 and PAM+7981 spacers were derived and the ‘priming protospacer’ that is targeted by s13-crRNA; thus, adaptation to this plasmid could be efficiently primed in DF60. A new-spacer-specific primer and s2-primer were together used to detect the acquisition of the PAM+7908 and PAM+7981 spacers. Lane M, dsDNA size marker. (C and D) The 3΄-end of the PAM+7908 and PAM+7981 spacers tended to change when their protospacer was mutated. The different nucleotide(s) between the wild-type (wt) pVS and its mutant (mut) p7908mut/p7981mut are framed. These plasmids were separately subjected to the adaptation assay illustrated in panel B, and the PCR products were subjected to high-throughput sequencing to characterize the spacer 3΄-end variation.