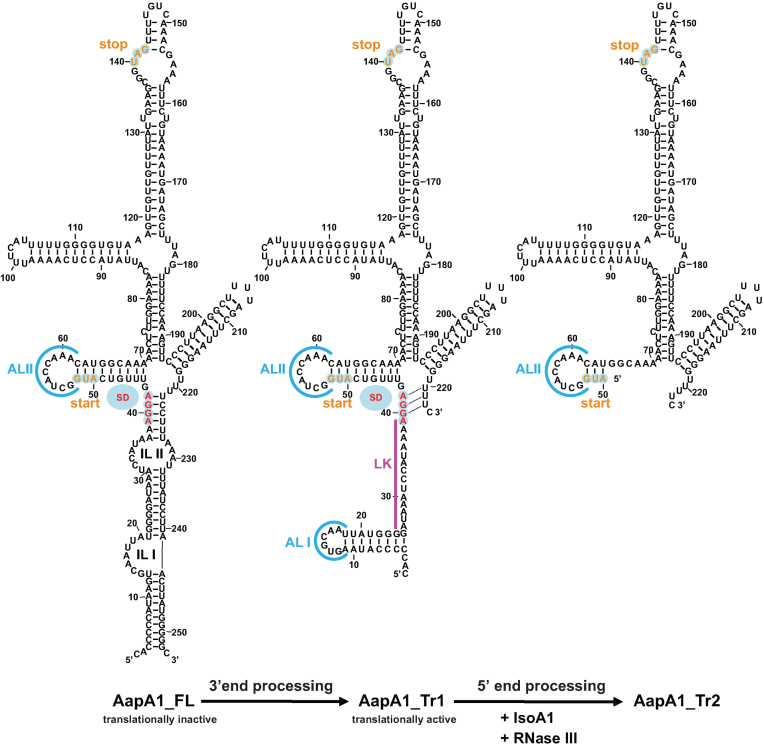
Figure 5.
mRNA folding predictions of the different AapA1 transcripts. The sequences of the three different AapA1 mRNA species were inferred from the RNA-seq data (4) and from Northern Blot analyzed with a combination of various probes (Supplementary Figure S3). Each secondary structure was predicted using RNAstructure 5.2 software (22) in agreement with the experimental data obtained by enzymatic and chemical footprinting (Figure 4 and Supplementary Figure S5). The secondary structures were visualized and designed with the VARNA applet (46). The AapA1-FL transcript corresponds to a 253 nt transcript in which most of the 5΄ UTR is engaged in a long distance interaction with the 3΄ end of the message. A 3΄ processing event removing the last 30 nt leads to the formation of a truncated transcript (AapA1_Tr1) that undergoes a structural rearrangement, rendering the SD accessible to the ribosome. The first 76 nt of this translationally active message interact with IsoA1 to form an extended duplex (not shown here, see Supplementary Figure S5). This long duplex is then cleaved by RNase III to generate the AapA1_Tr2 transcript. The start and stop codons of the AapA1 ORF are indicated in orange and the SD sequence is in red. Apical loops (AL) are shown in blue. Two internal loops (IL) are shown in the AapA1_FL transcript.