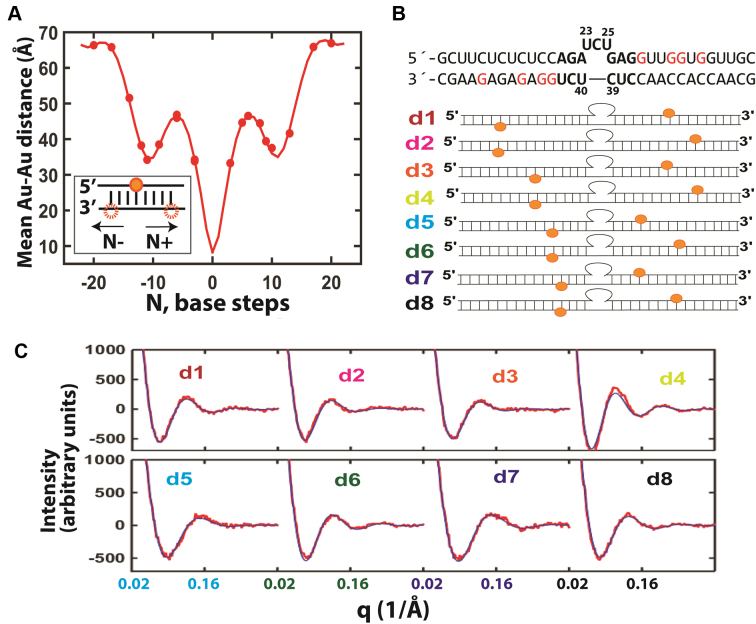
Figure 2.
Using XSI-RNA to resolve TAR RNA ensembles. (A) A solution model of RNA helices labelled with Au nanocrystals was developed from the average distances between two Au nanocrystals as a function of the number of separating base steps (30 mM Tris-HCl, pH 7.4, 150 mM NaCl and 10 mM sodium ascorbate; see Supplementary Figure S3 for RNA sequences). The data (circles) were fitted to a helical model of RNA (line; see Supplementary Data Notes S2–4 and Supplementary Table S1). The twenty sets of data points overdetermine the nine fitting parameters of the model. (B) The RNA constructs used for the TAR RNA XSI measurements. For each construct d1–d8 a pair of Au nanocrystals (orange) were attached to modified G residues (red). (C) The measured Au–Au scattering profiles (red) and the scattering profiles calculated from the resolved TAR ensemble (blue) for the constructs in part B (low salt condition: 30 mM Tris-HCl, pH 7.4, 10 mM NaCl and 10 mM sodium ascorbate; see Supplementary Figure S12 for higher salt profiles and Supplementary Figure S13 for the Au–Au distance distributions). An average value of χ2 = 1.5 was obtained for the fits.