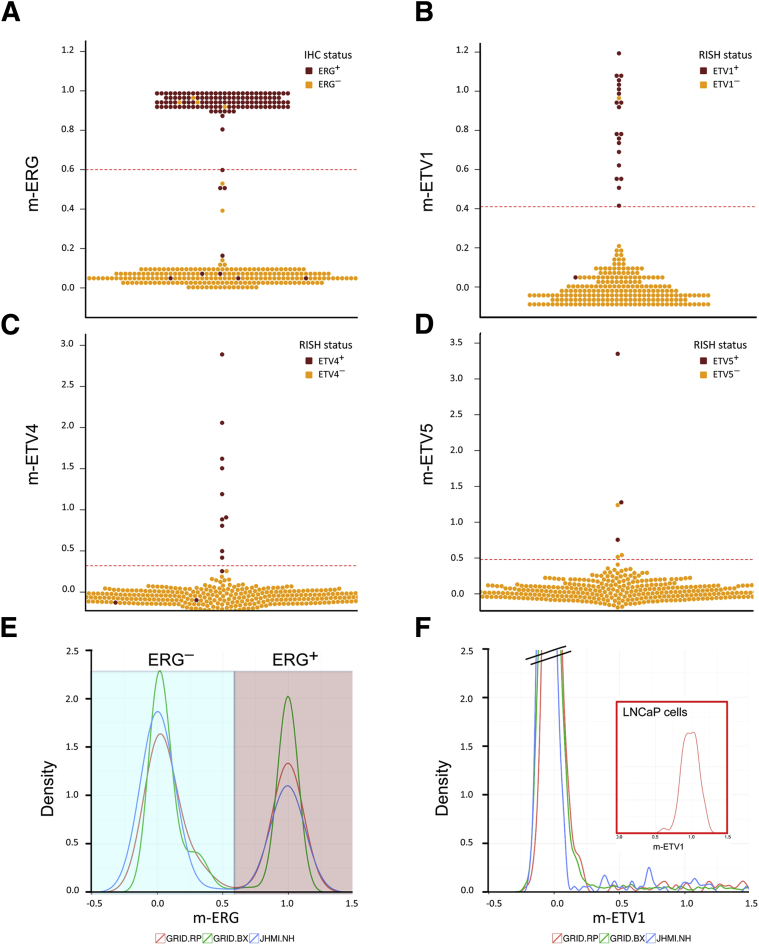
Figure 2.
Validation of the m-ERG and ETV1/4/5 gene expression models by comparison with IHC and RISH. A–D: Beeswarm plots of ERG, ETV1, ETV4, and ETV5 models in JHMI RP cohort showing model scores are highly concordant with IHC and RISH calls. The red dotted lines indicate the threshold of each model for calling ETS positivity. E and F: Density plots of m-ERG and ETV1 models in a subset of the JHMI RP cohort, prospective Decipher GRID RP, and biopsy cohorts and LNCaP cells with ETV1 fusion, showing distribution of models are highly similar across different data sets. ERG−, blue background; ERG+, mauve background. n = 358 JHMI RP cohorts (E and F); n = 4036 prospective Decipher GRID RP cohorts (E and F); n = 509 biopsy cohorts (E and F). GRID, Genomics Resource Information Database; IHC, immunohistochemistry; JHMI, Johns Hopkins Medical Institute; RISH, RNA in situ hybridization; RP, radical prostatectomy.