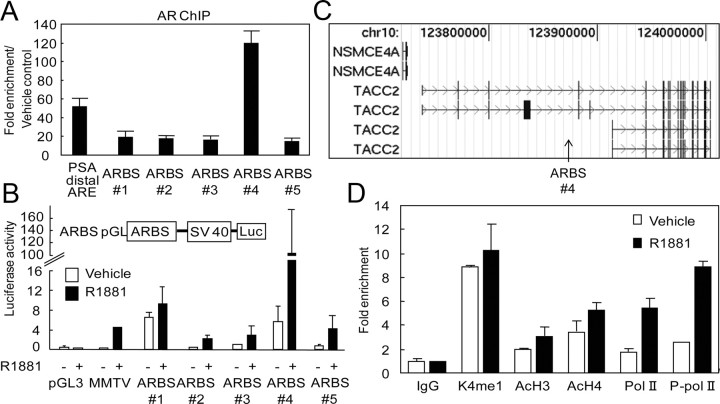
Fig. 1.
Characterization of the ARBS in the genomic region of the TACC2 gene. A, AR binding ability of the top five (ARBSs 1–5) of 100 ARBS determined by the ChIP-cloning strategy. LNCaP cells were treated with R1881 (10 nm) or vehicle for 24 h. The known distal ARE in the enhancer region of PSA was used as a positive control. Fold enrichment of AR binding was quantified by real-time PCR. The data represent the mean ± sd, n = 3. B, Androgen-dependent promoter activity of the top five ARBS evaluated by the luciferase reporter assay. LNCaP cells were transfected with pGL promoter vectors containing ARBS sequences 1–5. Empty pGL3 promoter and mouse mammary tumor virus (MMTV) luciferase vectors were used as negative and positive controls, respectively. Cells were treated with R1881 (10 nm) or vehicle for 24 h. The data represent the mean ± sd, n = 3. C, Location of ARBS 4 (arrow) in the TACC2 gene mapped by the UCSC Genome Browser (human genome NCBI Build 35). D, Active histone modification and RNA Pol II-dependent transcription at TACC2 ARBS. LNCaP cells were treated with R1881 (10 nm) or vehicle for 24 h. ChIP analysis was performed using nonspecific IgG or specific antibodies targeting histone H3 monomethylated K4 (K4me1), acetylated histone H3 (AcH3), acetylated histone H4 (AcH4), Pol II, or phosphorylated Pol II (P-pol II). Fold enrichment was quantified by real-time PCR. The data represent the mean ± sd, n = 2. Chr, Chromosome; SV40, simian virus 40.